Hello!
(1/7)
I'm thrilled to share my new article in @SoftXJournal !!
We present TFMLAB! An open source Matlab toolbox for 4D #TractionForceMicroscopy. We put special focus on making it accessible, even if you are not good at programming! 💻🧐
sciencedirect.com/science/articl…
🧵👇
(1/7)
I'm thrilled to share my new article in @SoftXJournal !!
We present TFMLAB! An open source Matlab toolbox for 4D #TractionForceMicroscopy. We put special focus on making it accessible, even if you are not good at programming! 💻🧐
sciencedirect.com/science/articl…
🧵👇
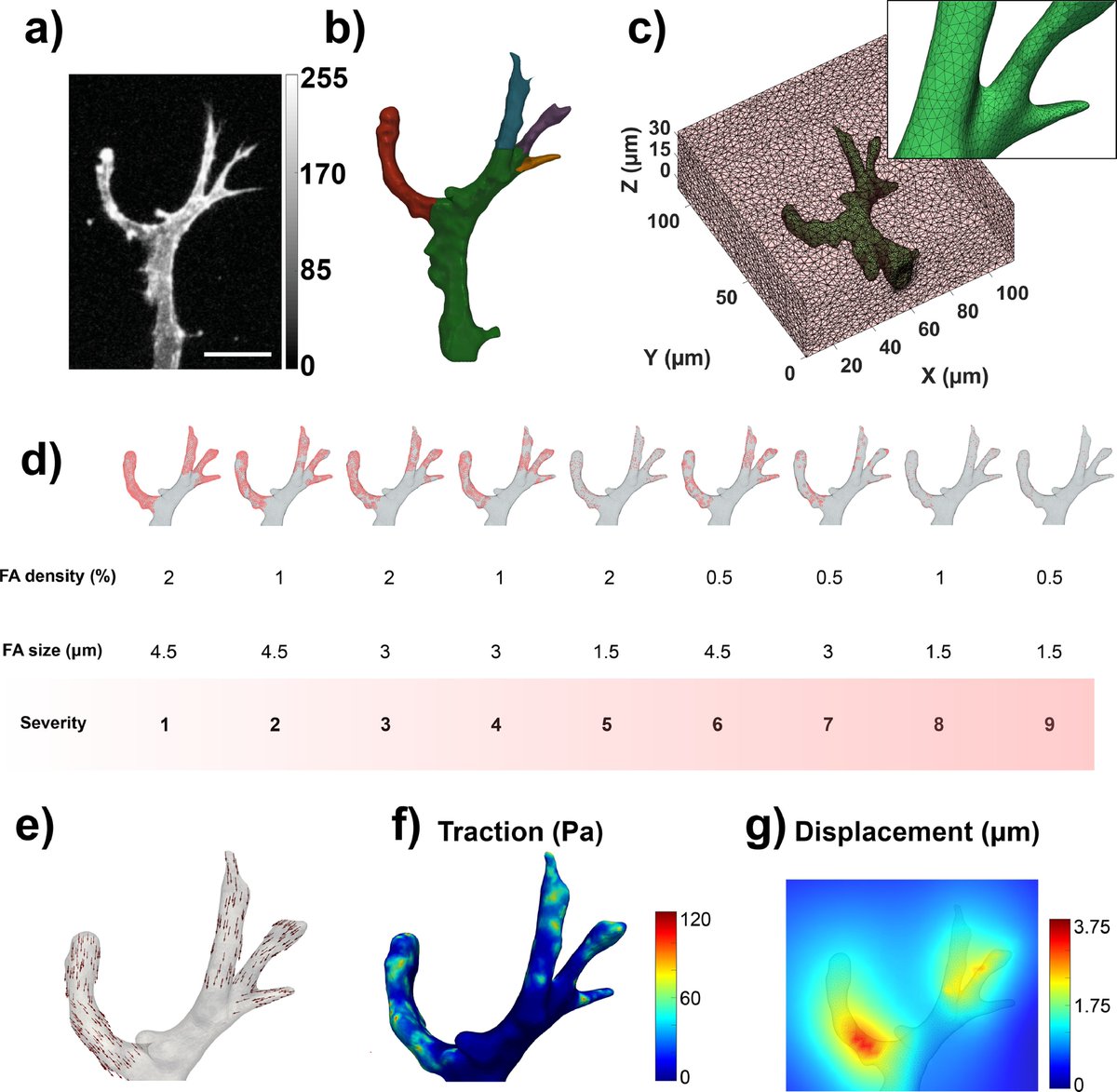
(2/7) There are many available open source codes to run 2D TFM, but what if you are now thinking of embedding your cells in a 3D matrix? You won't find that many #3DTFM codes out there that are easy to use without requiring you to be an expert programmer🧐💻
(3/7) Well, we have created #TFMLAB 🥳!! This Matlab toolbox integrates all the computational steps to compute active cellular forces from confocal microscopy images, including image processing, cell segmentation, matrix displacement measurement and force recovery 

(4/7) The good thing is that you can follow all these steps via graphical user interfaces which makes it easy to understand what is going on and to tune some important parameters! In the background, TFMLAB uses methods that we've developed over the last years in @MAtrix_KULeuven 

(5/7) As an example of application, we used TFMLAB to measure matrix displacements and tractions around angiogenic sprouts invading a #PEG hydrogel:
(6/7) TFMLAB provides output files and templates to visualize results in #Paraview. You don't need any licensed software to run the workflow other than Matlab. All the steps can be performed using open source software, which TFMLAB calls in the background! 

(7/7) Finally, I would like to thank all my collaborators in @MAtrix_KULeuven, @apek_shapeti, @MojBarz and prof. José Antonio Sanz from @unisevilla. We made it! #TFMLAB is finally public!! 🥳
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh