Is it possible to predict signaling in single cell? 🤔
Our @attila_gbr with Marco Tognetti, @BodenmillerLab group, @tanevski, and @Sagebio designed a @DR_E_A_M challenge to crowdsource this question and our manuscript is out 🎊biorxiv.org/content/10.110…. Here is what we learnt🧑🎓
Our @attila_gbr with Marco Tognetti, @BodenmillerLab group, @tanevski, and @Sagebio designed a @DR_E_A_M challenge to crowdsource this question and our manuscript is out 🎊biorxiv.org/content/10.110…. Here is what we learnt🧑🎓

we used this biorxiv.org/content/10.110… rich dataset from the @BodenmillerLab and @Picotti_Lab labs: 80 millions of #singlecell from 67 #breastcancer cell lines, 36 measured markers, 5 kinase inhibitors, time courses #CyTOF + #proteomics , transcriptomics and genomics 

Did the antibody stop working or you cannot measure a node? We can predict it’s activity from other measured nodes. Predictions correlate strongly with test data and are accurate across time and conditions. Watch out for rare signaling patterns though. 

Are you tight on budget or want to save time with a new inhibitor? Models can predict single cell response to new treatment. Predict individual protein activity AND interplay of nodes. Accuracy close to bio.replica. Predictions resemble inter- and intra cell line heterogeneity 
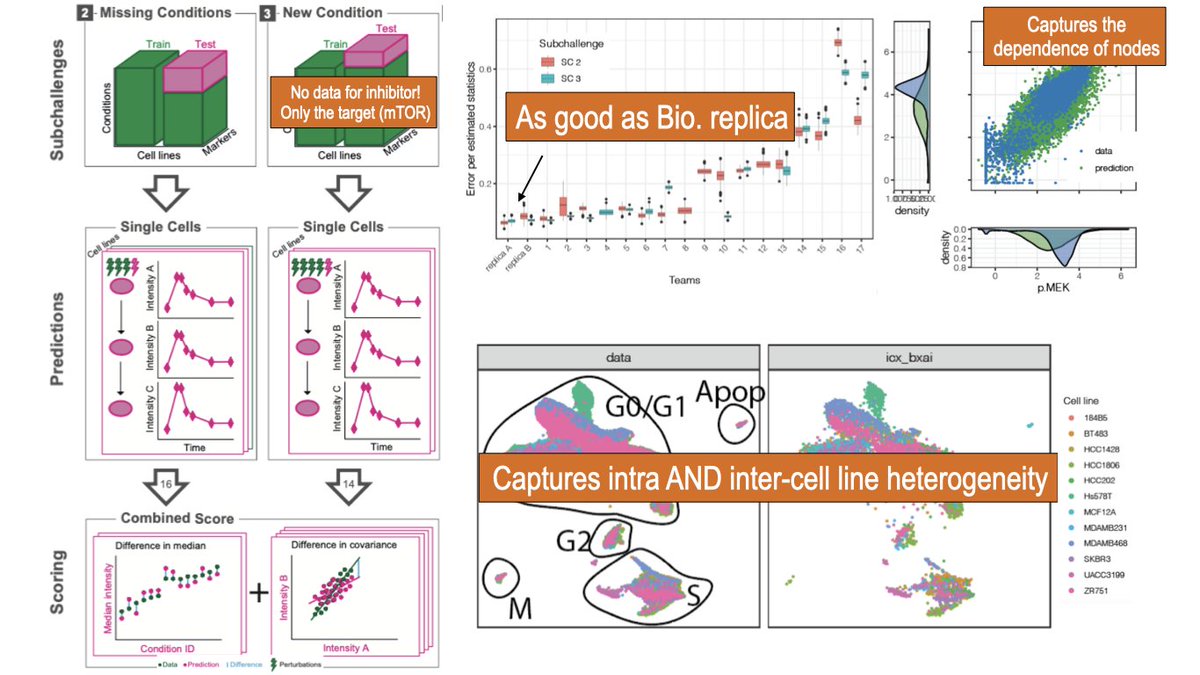
How much is dynamic response encoded in the basal activity and other omics state? More than we thought! Predictive models can reconstruct cell line specific dynamic response at population level. Accurate predictions across time and treatments especially for steady markers. 

Access all the data, all the submitted code and write ups at one place: synapse.org/singlecellprot…. Hope that best methods can be applied for new datasets and it will be a helpful reference for method developers. We thank all the challenge participants who made this possible! 👏
• • •
Missing some Tweet in this thread? You can try to
force a refresh