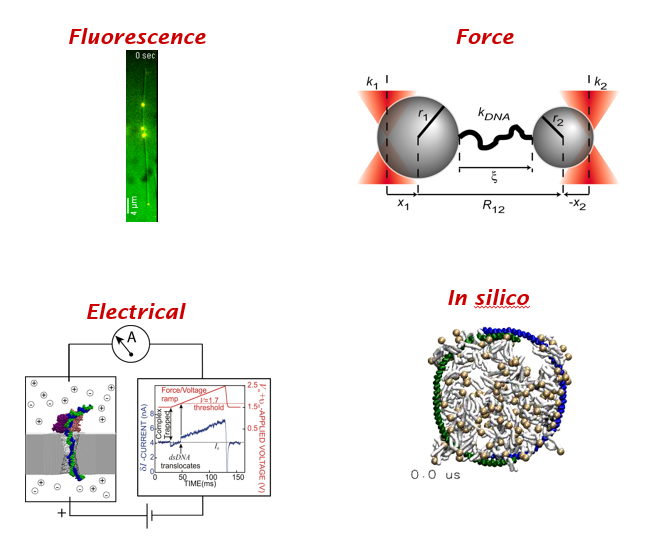
It's a beautiful Thursday! Time for a #DBIOtweetorial on bacterial cell division by @BioSciRy and @PetraLevin at the request of the awesome folks at #engageDBIO! 

On a first glance, bacterial cell division may seem simple. In reality, it is the culmination of precisely orchestrated interplay between cytoplasmic and extracellular processes. #EngageDBIO #DBIOTweetorial
To divide, bacteria must: grow, replicate and segregate their chromosome, add new cell wall perpendicular to the old cell wall, and separate. That’s a lot of work! #EngageDBIO #DBIOTweetorial 

One protein, FtsZ (in magenta below), is the star of bacterial cell division. FtsZ is a tubulin homologue that polymerizes in the presence of GTP to form a platform for the rest of the division machinery. Image courtesy of Jessie Bullock. #EngageDBIO #DBIOTweetorial 

Once the cell grows to the proper size and chromosome replication is complete, FtsZ polymerizes into short polymers around the inner circumference of the cell. These polymers treadmill, just like tubulin! #EngageDBIO #DBIOTweetorial 

FtsZ treadmilling ensures new cell wall is added evenly at the division site. This figure from Dr. @fleshball and colleagues shows old cell wall in blue and new cross wall labeled first in green and finally red. #EngageDBIO #DBIOTweetorial doi.org/10.1126/scienc… 

A cell has reached the right size for division when each relevant division protein is present in the correct *number* at the septum to proceed. Interestingly, this depends on the number of each protein present, not their cellular concentration. #EngageDBIO #DBIOTweetorial 

Rather than being directed to the future division site by a mark of some kind, in many bacteria FtsZ is kept *away* from the cell poles and the chromosome by numerous factors that corral the division protein to midcell. #EngageDBIO #DBIOTweetorial 

Some aspects of cell division remain unclear. For example, the mechanisms ensuring that the gram-negative outer membrane, which is external to the newly made cross wall, divides along with the rest of cell envelope are not entirely clear. #EngageDBIO #DBIOTweetorial 

Accordingly, bacterial cell division is an active field. Some of the folks to follow here on twitter are: @JoeLutkenhaus @goleylab @jiexiao_lab @CWoldringh @_Alex_Bisson_ @junlab_ucsd @ProfLizHarry @GillesvanWezel @seamus_holden @ajlabny @nartimsoole #EngageDBIO #DBIOTweetorial 

• • •
Missing some Tweet in this thread? You can try to
force a refresh