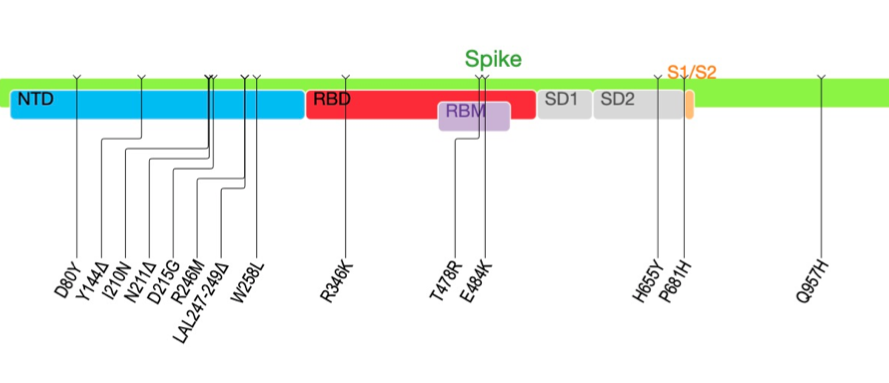
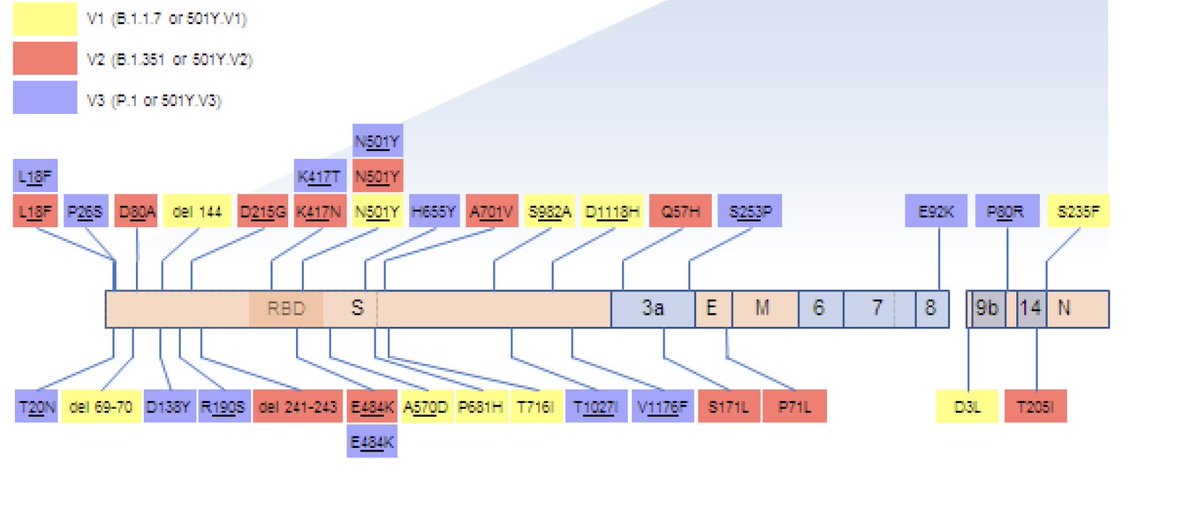
First genomic surveillance results from a traveler to India in South Africa. A patient at Tygerberg Hospital who tested positive for SARS-CoV-2 about 3 days after traveling to SA from India. This first genome is a 501Y.V2 (B.1.351) and not the B.1.617 described in India.
The 501Y.V2 (B.1.351) is the 3rd most common variant in India after the B.1.617 and the B.1.1.7. Below a graph from @trvrb showing the spread of the variants in India. 

The 501Y.V2 (B.1.351) was firstly identified in South Africa by the network for genomic surveillance in South Africa (NGS-SA). It shows how variants can be introduced back and forth between countries.
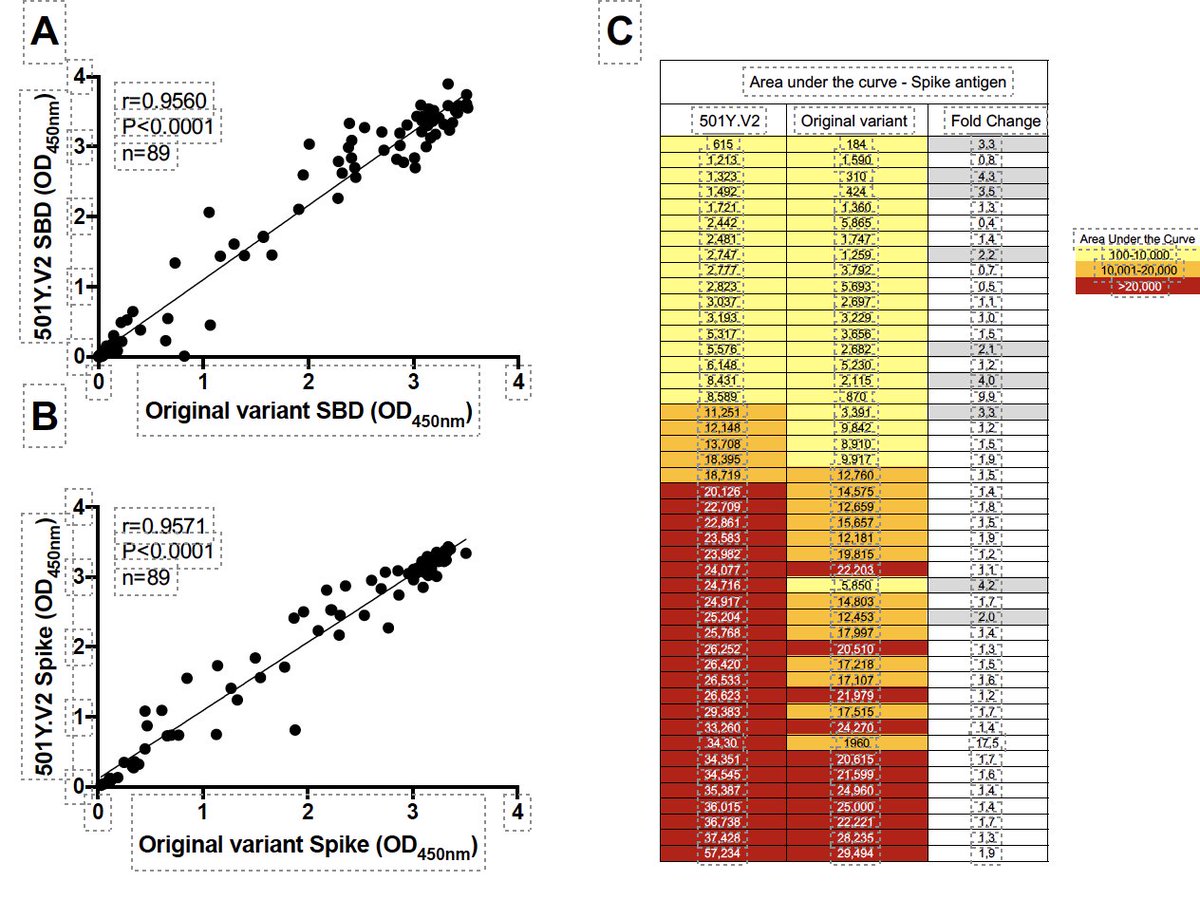
At present, we believe that the 501Y.V2 (B.1.351) antibody response may be strong enough to neutralize the B.1.617 as the 501Y.V2 can neutralize other lineages and variants with many mutations, such as P1 - see some of SA papers at: nejm.org/doi/full/10.10…
Another important paper showing the neutralization breath of the 501Y.V2 on the paper at: nature.com/articles/s4158…
The identification of the first genome from a traveller from India in South Africa was due to very diligent and fantastic work of Prof Susan Engelbrecht, who is a member from the Network for Genomic Surveillance in South Africa (NGS-SA) from @StellenboschUni
We are strengthening genomic surveillance in South Africa. So far, the dominant variant is the 501Y.V2 (around 99%), we have also detected multiple introductions of B.1.1.7 and the A.23.1. We will keep reporting on results and keep an eye for new variants.
The very important take home message is that the known public health interventions (testing, contact tracing and isolation) together with non-pharmaceutical interventions (social distance, washing hands and wearing masks) are extremely important as we scale vaccination.
• • •
Missing some Tweet in this thread? You can try to
force a refresh