A year of genomic surveillance reveals how the SARS-CoV-2 pandemic unfolded in Africa medrxiv.org/content/10.110…
By 5 May 2021, 14,504 SARS-CoV-2 genomes had been submitted to the GISAID database from 38 African countries and two overseas territories (Mayotte and Réunion) (Fig. 1A). Overall, this corresponds to approximately one sequence per ~300 reported cases. 

Overall, the number of sequences correlates closely with the number of reported cases per country. lmost half of the sequences were from South Africa, consistent with it being responsible for almost half of the reported cases in Africa. 

The countries/territories with the highest coverage of sequencing (defined as genomes per reported case) are Kenya (n=856, one sequence per ~203 cases), Mayotte (n=721; one sequence per ~21 cases), and Nigeria (n=660, one sequence per ~250 cases).
Half of all African genomes were deposited in the first ten weeks of 2021, suggesting intensified surveillance in the second wave following the detection of B.1.351/501Y.V2 and other variants
In total, we detected at least 730 viral introductions into African countries between February 2020 and February 2021, over half of which occurred before the end of May 2020.
Whilst the early phase of the pandemic was dominated by importations from outside Africa, predominantly from Europe. 

There was then a shift in the dynamics, with an increasing number of importations from other African countries as the pandemic progressed. 

South Africa, Kenya and Nigeria appear as major sources of importations into other African countries. Particularly striking is the southern African region, where South Africa is the source for a large proportion (~80%) of the importations to other countries in the region. 

Africa has also contributed to the international spread of the virus with at least 356 exportation events from Africa to the rest of the world. Consistent with the source of importations, most exports were to Europe (41%), Asia (26%) and North America (14%).
An increase in the number of exportation events occurred between December 2020 and March 2021, which coincided with the second wave of infections in Africa and with some relaxations of travel restrictions around the world.
After its emergence in South Africa, B.1.351 became the most frequently detected SARS-CoV-2 lineage found in Africa. It was first sampled on 8 October 2020 in South Africa (13) and has since spread to 20 other African countries. 

Although lineage A viruses were imported into several African countries, they only account for 1.3% of genomes sampled in Africa. Lineage A is the oldest SARS-CoV-2 lineage, representing the original Wuhan isolates of the virus from December 2019
there is evidence of an increasing prevalence of lineage A viruses in some African countries. In particular, A.23.1 emerged in East Africa and appears to be increasing rapidly in prevalence in Uganda and Rwanda. 

Furthermore, a highly divergent variant from lineage A was recently identified in Angola from individuals arriving from Tanzania - medrxiv.org/content/10.110…
n order to determine how some of the key SARS-CoV-2 variants are spreading within Africa, we performed phylogeographic analyses on the VOC B.1.351, the VOI B.1.525, and on two additional variants that emerged and that we designated as VOIs for this analysis (A.23.1 and C.1.1). 

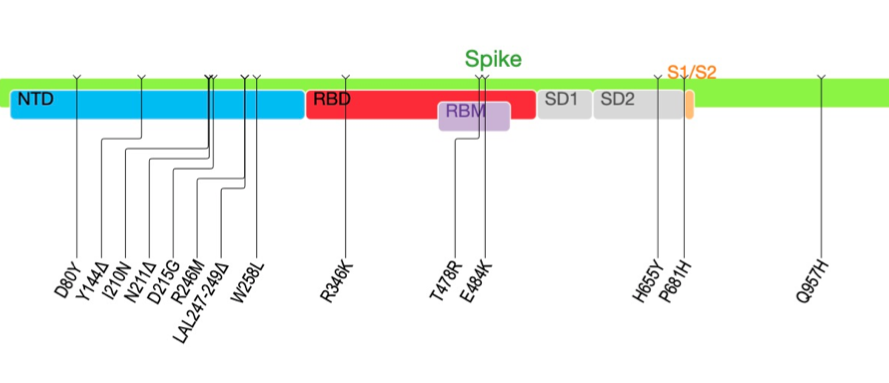
These African VOCs and VOIs have multiple mutations on Spike glycoprotein and molecular clock analysis of these four datasets provided strong evidence that these four lineages are evolving in a clocklike manner. 

The B.1.525 is spreading widely in West Africa and C.1., which has originated in South Africa have added new Spike mutations are spreading north and east in the south of the continent 

Conclusions: Although distorted by low sampling numbers and blind-spots, the findings highlight that Africa must not be left behind in the global pandemic response, otherwise it could become a breeding ground for new variants.
There are tons of people to thank! Over 30 countries, @GISAID, >100 organizations and many of the great funders in the world that believe that Africa can lead in their response to pandemics!
Paper open accessible at: medrxiv.org/content/10.110…
• • •
Missing some Tweet in this thread? You can try to
force a refresh