
Our preprint about reinfection by the P.1 SARS-CoV-2 lineage in repeat blood donors in Manaus is now out [not peer-reviewed] 🧪🦠🥼🌎
▶️Check it out at: medrxiv.org/content/10.110….
▶️Short thread on key findings🧵 1/10
▶️Check it out at: medrxiv.org/content/10.110….
▶️Short thread on key findings🧵 1/10
It is challenging to estimate the reinfection rate since most reinfected individuals are likely not tested, and/or oligo or asymptomatic. Yet, reinfection has been postulated to explain at least in part the 2nd wave of COVID-19 in Manaus (see bit.ly/3hyjVWk). 2/10
To address this, we tested serial samples from unvaccinated repeat blood donors from Manaus w a seroprevalence assay that wanes during convalescence. We assumed that reinfections induce a boosting ↗️ in antibody (Ab) levels, leading to V-shaped time series of Ab reactivity. 3/10
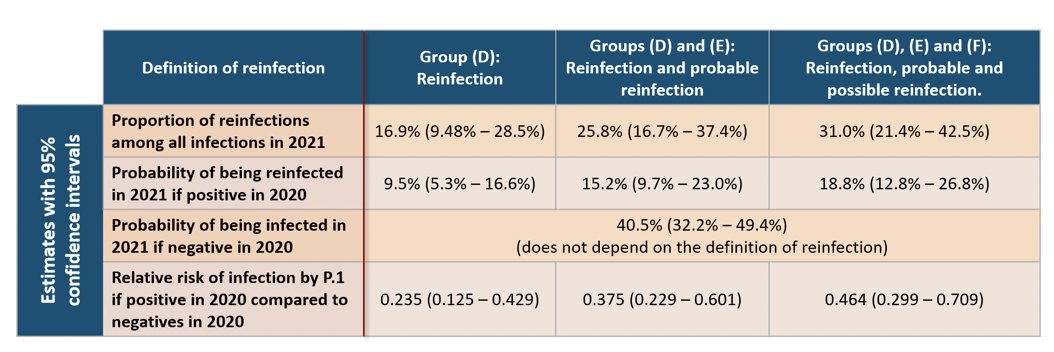
We selected 238 repeat donors based on their date of donation and partitioned them into 6 groups. These groups reflect the inferred sequence of infection and reinfection with non-P.1 and P.1 variants. 4/10 

The Ab signal-to-cutoff (S/C) curves for the 6 groups (A–E) are shown below. Blue dots are positive donations (S/C>=0.49), red dots are negative donations, and donations of the same donors are connected by a line. 5/10 

Cases of probable or possible reinfections (defined
by considering the time period when the antibody levels are expected to grow after recovery and
the range of half-lives for antibody waning after seroconversion) were also considered. 6/10
by considering the time period when the antibody levels are expected to grow after recovery and
the range of half-lives for antibody waning after seroconversion) were also considered. 6/10

P.1 dominated in Manaus from Jan 2021 onwards (see bit.ly/3tRYfXz). Based on our classification criteria, we infer that of the presumed P.1 infections observed in 2021, between 16.9 (9.5%–28.5%) to 31.0% (21.4-42.5%) were reinfections. 7/10 
There are several limitations of the study. For example, repeat negative donors may not represent truly unexposed individuals because sparse sampling may have resulted in missing the interval with a positive S/C signal. 8/10
Moreover, blood donors are biased towards asymptomatic and mild infections; therefore, our rates of reinfection cannot be extrapolated to persons with more severe primary infections. Despite these caveats 9/10
Our study suggests that reinfection due to P.1 is more frequent than what is typically detected by traditional epi, molecular and genomic surveillance of clinical cases. 10/10
This work was led by @CarlosPrete1 @LewisBuss4 Claudia Abrahim Tassila Salomon @Myuki97859555 Marcio Oikawa Renata Buccheri Eduard Grebe Allyson da Costa Nelson Fraiji Maria Carvalho Neal Alexander @nmrfaria @chris0dye Vitor Nascimento Michael Busch @estercsabino
And we come from a variety of institutions @imtspusp @usponline @fhemoam @hemominas @ufabc Vitalant Research Institute @UCSF #SACEMA @StellenboschUni @ImperialSPH @MRC_Outbreak @UniofOxford @OxZooDept
Many thanks to all study participants and to our funders @The_MRC @AgencyFAPESP @wellcometrust @FMUSPoficial @CNPq_Oficial @nih_nhlbi @itau #TodosPelaSaude
• • •
Missing some Tweet in this thread? You can try to
force a refresh





