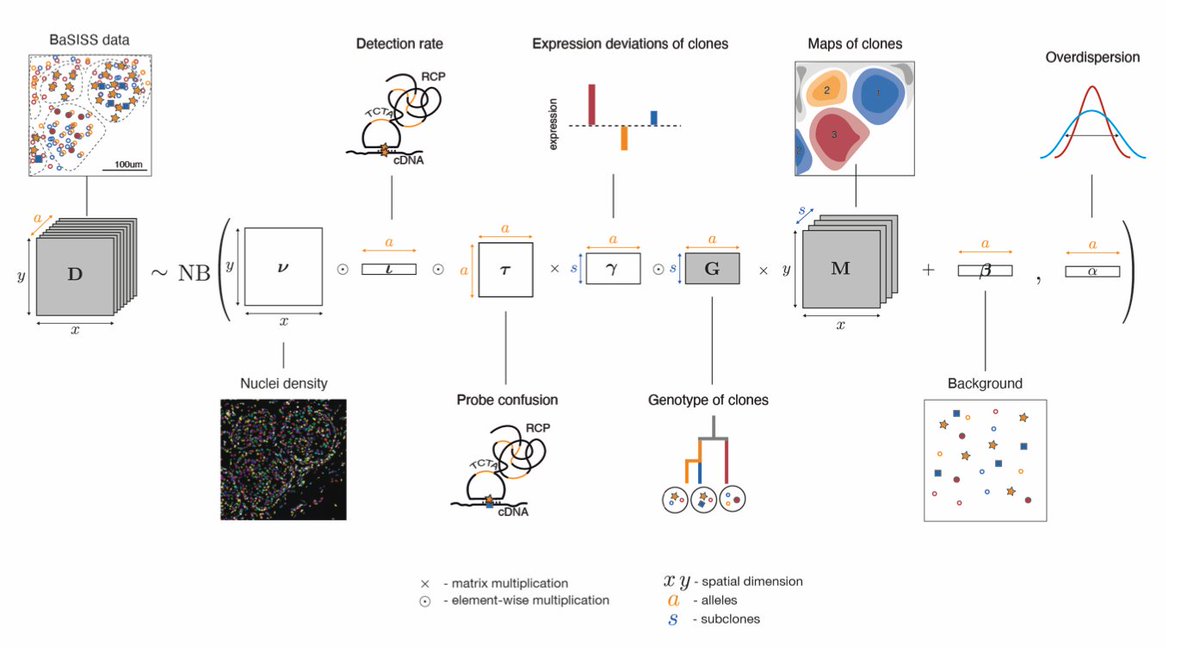
What have we learned from analysing 200,000 SARS-CoV-2 genomes from genomic surveillance in England in the last 9 months?
These data provide important context for the current situation related to B.1.617.2.
Here’s a summary of the preprint. medrxiv.org/content/10.110…
These data provide important context for the current situation related to B.1.617.2.
Here’s a summary of the preprint. medrxiv.org/content/10.110…

@harald_voeh has developed a model that tracks 62 different lineages across 315 local authorities in England. His model estimates total and lineage-specific incidence and growth rates. 

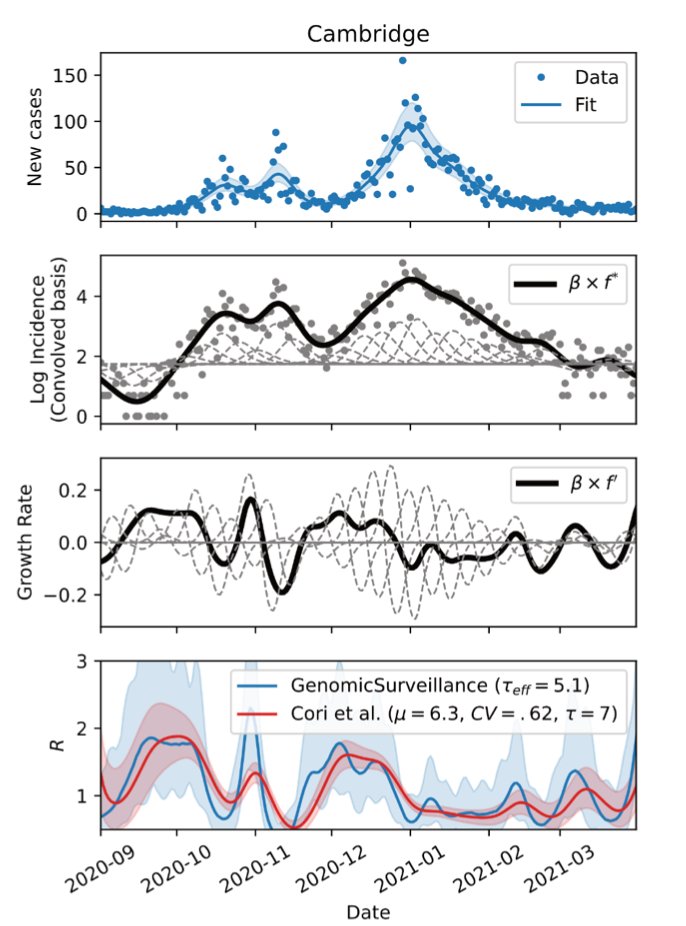

The model also calculates lineage-specific relative growth rates and provides a fairly accurate reconstruction of the epidemic and its many subepidemics across the nation between Sep '20 and Apr' 21. We also included a provisional analysis until 15 May '21 to track B.1.617.2 
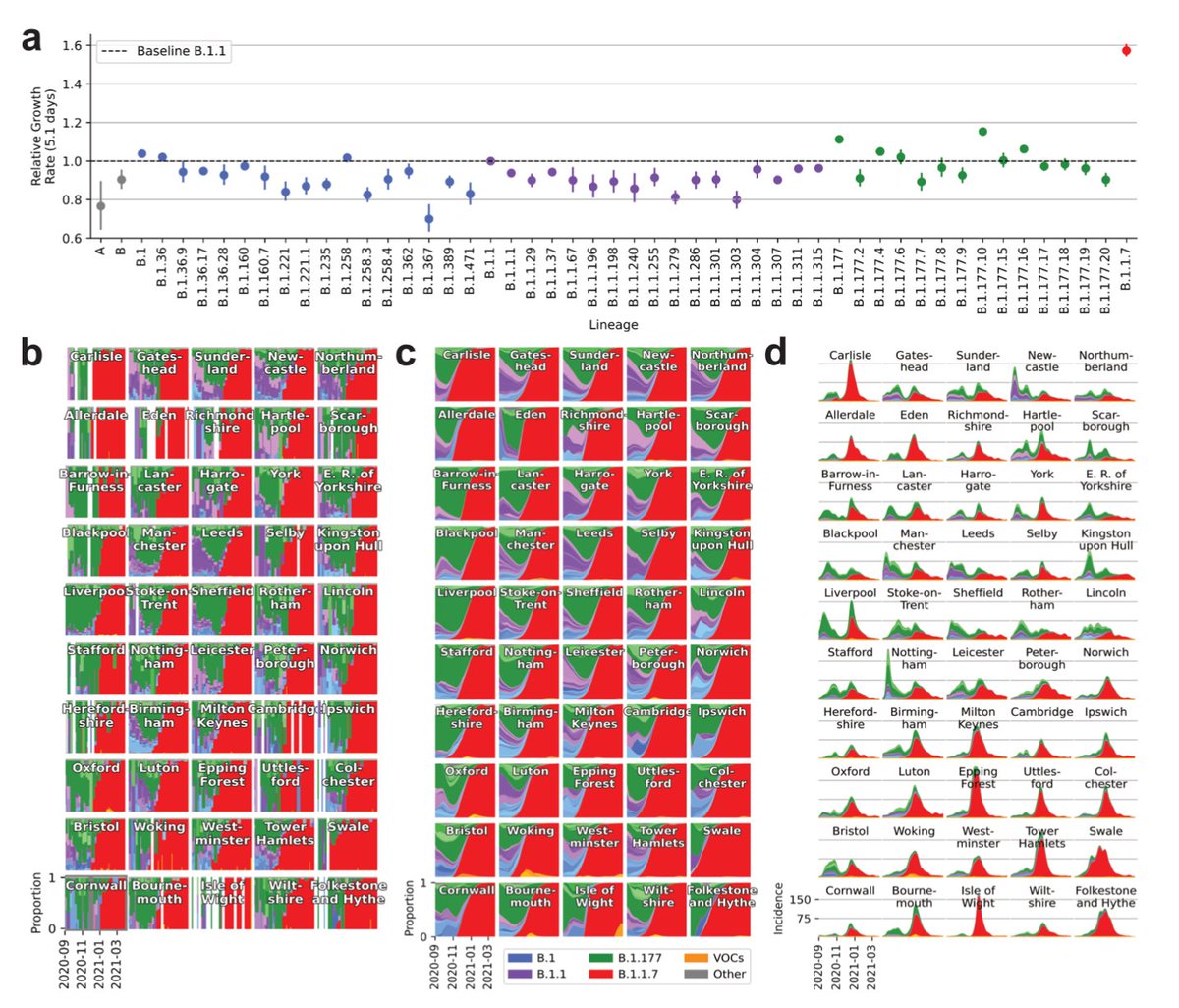
This allowed us to take a closer look at how B.1.1.7 spread during periods of lockdown and regionally tiered restrictions from Nov to Dec '20. The geographic and temporal correspondence between growth rates and restrictions is striking. Still, B.1.1.7 was simply too fast. 

It was only the third national lockdown from Jan to Mar ‘21 (and an element of acquired and vaccine derived immunity) that stopped B.1.1.7.
Yet as a byproduct of the strong restrictions, almost all lineages present in Sep ‘20 were eliminated.
But some variants resisted.
Yet as a byproduct of the strong restrictions, almost all lineages present in Sep ‘20 were eliminated.
But some variants resisted.

Different E484K containing variants were repeatedly introduced (or emerged domestically) between Dec '20 and Mar ‘21. Yet they mostly caused short lived regional or local outbreaks and increased only very moderately over all. 



But the situation changed in Apr ‘21 when B.1.617.2 was introduced and spread rapidly, reaching a national frequency of 40% on May 15 ‘21. 



It was associated with a number of local outbreaks as in Bolton, but also spread to 227 LTLAs. We estimate it grew around 30-50% faster than B.1.1.7. since its introduction. 



We don’t understand exactly why B.1.617.2 spread so rapidly. Three factors could have contributed: Transmissibility and/or immune (vaccine) escape, repeated introductions and demographic factors facilitating onwards transmission.
Regarding introductions and demographic factors it is instructive to study its sister lineage B.1.617.1, which would largely share these factors, but didn’t grow much and produced much smaller clades per introduction. 



Recent data by PHE and other investigators points towards higher secondary attack rates and reduced vaccine efficiency, especially for single doses, yet some questions remain as summarised by co-lead @jcbarett
https://twitter.com/jcbarret/status/1396913527110123523?s=20
So far B.1.617.2’s relative increase was a combination of its own growth and B.1.1.7’s decline. But the equation might change in the future: Outbreaks may cede, circulation in the wider population increases, the effects of reopening and also further vaccination.
As we don’t have all the answers yet it becomes clear that further and rapid genomic surveillance is critical. @theosanderson and others have developed a website where these data can be explored. covid19.sanger.ac.uk/lineages/raw
A great thank you to everyone who has contributed to this work - this was an extremely collaborative project that couldn’t have been realised by any single individual. 🙏 

• • •
Missing some Tweet in this thread? You can try to
force a refresh