In this online pre-print, we present a case of persistent SARS-CoV-2 infection with accelerated intra-host evolution in a patient with advanced HIV and antiretroviral treatment failure in South Africa.
Paper deposited at medRxiv &available at: krisp.org.za/publications.p… @sigallab
Paper deposited at medRxiv &available at: krisp.org.za/publications.p… @sigallab
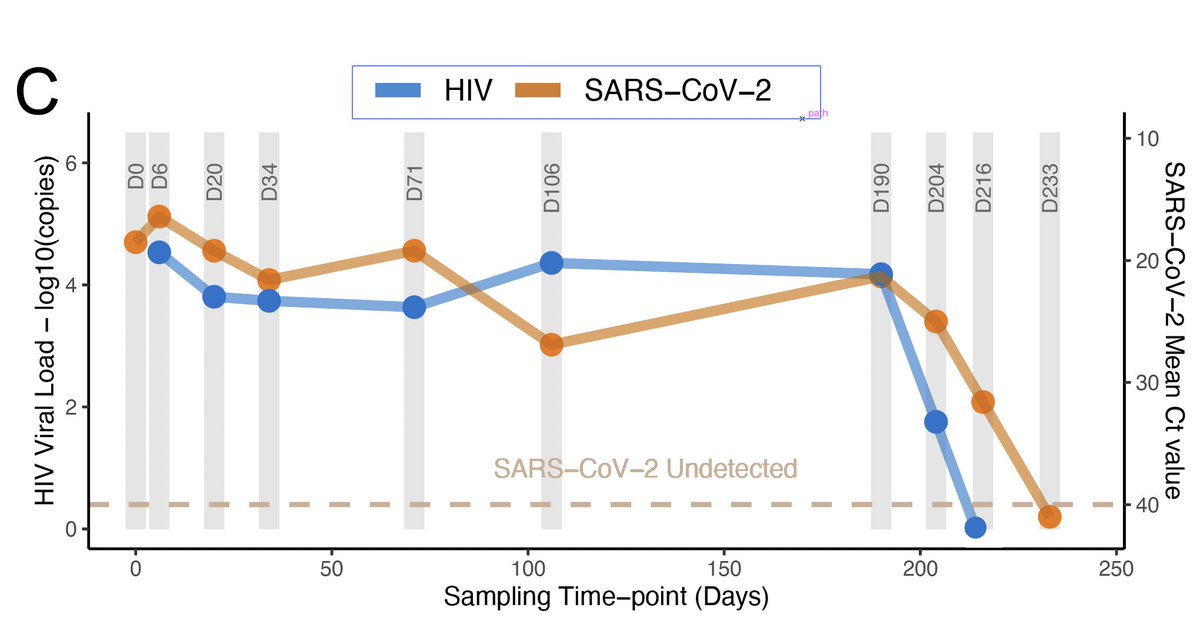
SARS-CoV-2 RT-PCR was positive for 216 days (27 weeks). At all time-points between day 0 and day 216 there was a mean Ct ranging from 16.4 to 31.6. The patient had HIV drug resistance and was not responding to therapy. 
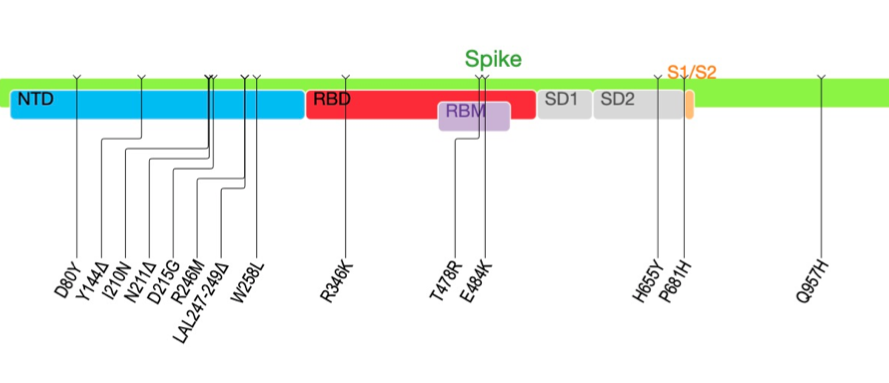
Over 30 mutations accumulated over time, many of them changing aminoacids that are common of variants of concern. 

At day 6, the E484K substitution in the receptor-binding domain (RBD) emerged, along with a substitution in the S2 subunit (A1078V). The E484K mutation remained in the consensus sequence until day 34, but then persisted at low frequency up to day 190
At the later time points, there was a significant shift in the virus population, with the emergence at day 71 of the K417T and F490S mutations in the spike RBD, followed by the emergence of L455F and F456L at day 106. At day 190, there was the emergence of D427Y and N501Y.
Table of mutations in the Spike protein, including the appearance of 501Y, 484K and 417T (all from variants of concern) and mutations and deletion at the N-terminal domain (NTD) 

All genomes fell within PANGO lineage B.1.1.2732, a South African lineage that was previously part of B.1.1.56, one of the most prevalent lineages in the first wave of the epidemic in the province of KwaZulu-Natal in South Africa. 

Discussion: Despite a short clinical illness of moderate severity, SARS-CoV-2 PCR positivity persisted up to 216 days. We demonstrate significant shifts in the virus population over that time, involving multiple mutations at key neutralizing antibody sites in the spike RBD & NTD
Most other cases of persistent infection have been described in people with haematological malignancies or people receiving immunosuppressive therapies for solid organ transplants or other chronic medical conditions
There is one other documented case of prolonged infection in a person with HIV - in that case the person was ART-naïve with profound immunosuppression and genome sequencing revealed only a single emergent mutation in spike (T719I) at day 53
The genomic and clinical data suggest that virus evolution may have been driven by selective pressure from an impaired neutralizing antibody response due to advanced HIV disease.
Disruptions to HIV programs as a result of COVID-19 could potentially increase the burden of advanced HIV45. These findings highlight the importance of maintaining essential health services in high HIV prevalence settings, and accelerating progress towards 90-90-90 targets.
If persistent SARS-CoV-2 infection and evolution of virus does occur more frequently in the context of HIV, it may provide justification for prioritising people living with HIV for COVID-19 vaccination.
Real teamwork with @farinakarim @sigallab @rjlessells @AHRI_News @krisp_news and the clinical team of the Infectious Diseases Department of UKZN!
• • •
Missing some Tweet in this thread? You can try to
force a refresh