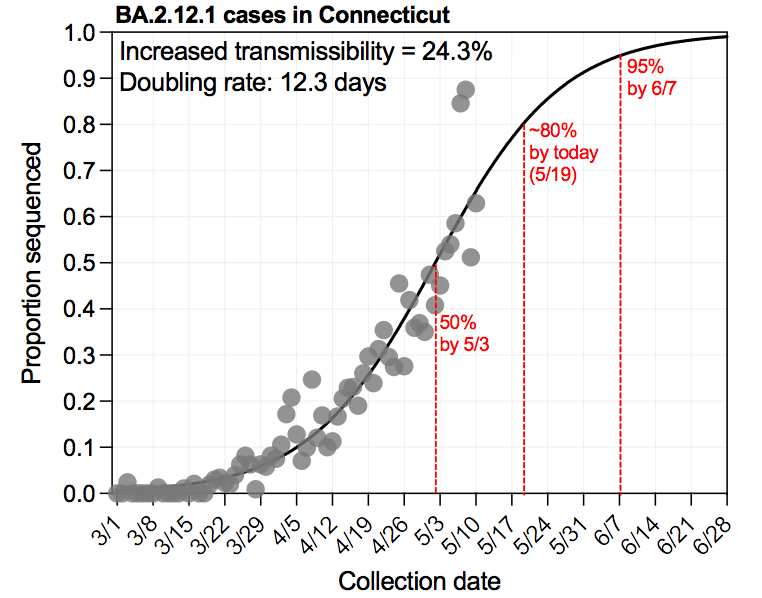
🧬6/10 Connecticut #SARSCoV2 variant surveillance
@CovidCT | @jacksonlab | @CTDPH | @YaleSPH | @Yalemed | @YNHH
Alpha (B.1.1.7) = 57% (📉11%)
Gamma (P.1) = 10.9% (📈2.7%)
Delta (B.1.617.2) = 6.5% (📈3.2%)
short 🧵 | Report 👉
covidtrackerct.com/variant-survei…
@CovidCT | @jacksonlab | @CTDPH | @YaleSPH | @Yalemed | @YNHH
Alpha (B.1.1.7) = 57% (📉11%)
Gamma (P.1) = 10.9% (📈2.7%)
Delta (B.1.617.2) = 6.5% (📈3.2%)
short 🧵 | Report 👉
covidtrackerct.com/variant-survei…
2/7 Gamma (P.1) and Delta (B.1.617.2) continue rising in Connecticut, while Alpha (B.1.1.7) and others decline, following national trends (see next tweet). 

3/7 Data from outbreak.info shows that in the US 🇺🇸, Delta (B.1.617.2) is 📈 exponentially, while Alpha (B.1.1.7) is on the 📉. Despite this, COVID-19 cases are still dropping (for now). 

4/7 Compare this to the UK 🇬🇧, where Delta (B.1.617.2) is dominant and Alpha (B.1.1.7) is approaching (but probably wont reach) extinction. And this is being following by a recent 📈 in cases. 

5/7 What will happen in CT and the 🇺🇸? Delta will likely become dominant. The 🇺🇸 and 🇬🇧 have quite similar 💉 rates, so we may even see an 📈 in cases like the 🇬🇧. But full vaxx can still be effective against Delta, so I'm guessing that it would be a large '🌊'.
(data from NYT)
(data from NYT)

6/7 How we prevent Delta (and other variants) from causing another 🌊 is pretty simple. Get vaccinated 💉.
7/7 This week I'd like to thank Kaya Bilguvar and our partners at YCGA who stood up COVIDseq and significantly increased our capacity. The entire team has been amazing to work with, and we could not have done this 👇 without them 👏🙌
medicine.yale.edu/keck/ycga/
medicine.yale.edu/keck/ycga/
**would NOT be a large wave. ugh.
• • •
Missing some Tweet in this thread? You can try to
force a refresh