New paper in @NatureMedicine on #mtDNA variant effects on metabolites, from my colleagues and I from @sangerinstitute, @emblebi , @Cambridge_Uni and @MRC_MBU! nature.com/articles/s4159…
@nicolesoranzo @cam_mito @OliverStegle @auroradevea @joannahowson @DoingYonova
🧵 1/n
@nicolesoranzo @cam_mito @OliverStegle @auroradevea @joannahowson @DoingYonova
🧵 1/n
We identified common #mtDNA variants in 16K British individuals with EUR ancestry from the INTERVAL study, performed the first large-scale assoc study between #mtDNA variants and ~5000 molecular traits that directly tag metabolic processes. 2/n 

We found and replicated significant associations between #mtDNA variants in Haplogroups Uk (found in 10% of the EUR population) and H3 (2%) and levels of N-formylmethionine (fMet), an amino acid known to be important for intra-mitochondrial translation. 3/n 

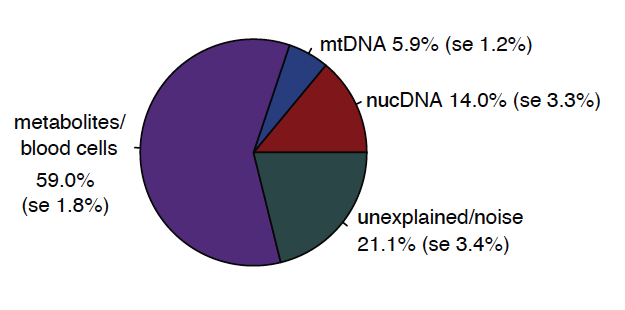
These #mtDNA effects are independent on nuclear DNA effects on levels of fMet, and account for 5.9% of the variation in fMet levels between individuals, a high percentage, for such a small (16KB genome)! 4/n 
As #mtDNA Haplogroup Uk has previously been shown to be protective against diseases such as #Parkinson’s (PD) and ischaemic stroke (IS), we investigated the molecular mechanisms that may mediate these protective effects. 5/n
We first show that #mtDNA variants in Haplogroup Uk regulate the expression of many #mtDNA genes encoding for proteins in the mitochondrial complexes, in a majority of tissues in the GTEx dataset. For eg, this figure shows effects of fMet associated variants on MT-ND3. 6/n 

We then show using extensive experiments on cybrid cell lines (same nuclear DNA, different #mtDNA) that #mtDNA variants in Haplogroup Uk regulate protein synthesis and degradation in both mitochondria and cytoplasm, and this affects cellular processes beyond #bioenergetics. 7/n 

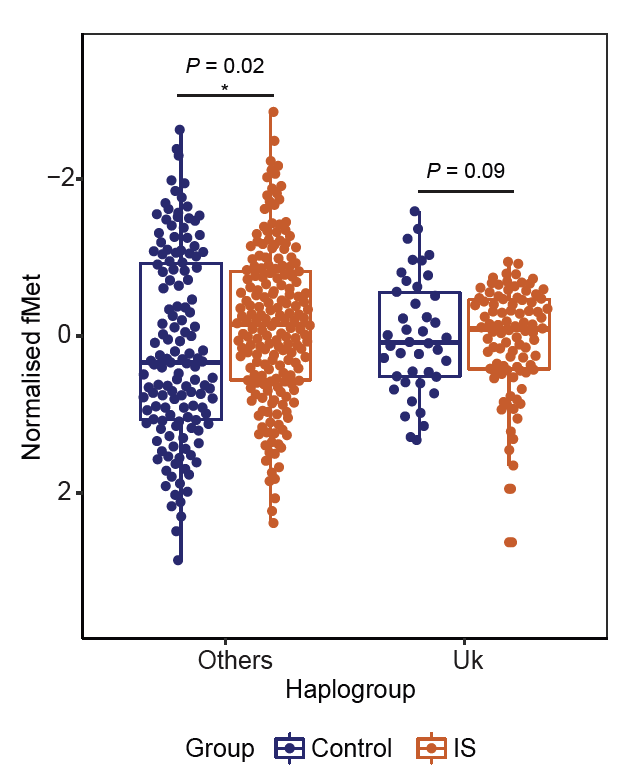
As for whether and how increased fMet in Haplogroup Uk may mediate protective effects against diseases: First, we measured fMet in a clinical cohort of IS, and found part of the protective effect of Haplogroup Uk on IS may be attributed to fMet. 8/n 
Second, we assessed the effects of fMet on the incidence of range of late-onset diseases in a prospective cohort with a 20-year follow-up EPIC-Norfolk, and found that fMet is associated with a wide range of diseases, and a potential biomarker for ageing and disease. 9/n 

In summary, our went from quantitative genetics ➡️ cellular models ➡️ clinical cohorts ➡️ prospective study, and sheds new light on unknown mechanisms through which #mtDNA variation contributes to metabolic processes, protein proteostasis, and liability to complex diseases. 10/n
Our results support the need for broad and hypothesis-free investigations on the impact of #mtDNA variation and nuclear-mitochondrial interactions on ageing and disease. With greater sample size and power, we will be sure to discover more. 🧐11/n
This is truly to a team effort, and a great example of how interdisciplinary collaboration can lead to real insight!! ❤️❤️❤️
@aidanbutty @Claudia20755263 @pietznerm @kousikbioinfo @mjbonder and many many others :)
@aidanbutty @Claudia20755263 @pietznerm @kousikbioinfo @mjbonder and many many others :)
• • •
Missing some Tweet in this thread? You can try to
force a refresh