
Earlier this week we put out a couple of preprints (doi.org/10.1101/2021.0… doi.org/10.1101/2021.0…) describing minoTour github.com/LooseLab/minot…, an @nanopore tool we have been developing for several years. A bit of a 🧵
The minoTour design philosophy is all around real time - it's about working out what is happening at that moment - with the exception of ARTIC (see below) we anticipate users running further analysis after minoTour!
What is minoTour? It's a real time platform for monitoring and analysing nanopore data. We use it routinely in the lab to track everything we sequence (and flow cell QC too!). It's main day-to-day use is just keeping track of whats happening. 

We can monitor the sequencer directly ("Live Data") and get instant remote feedback on how well a run is working and even send messages back to MinKNOW and stop runs once they are finished. 



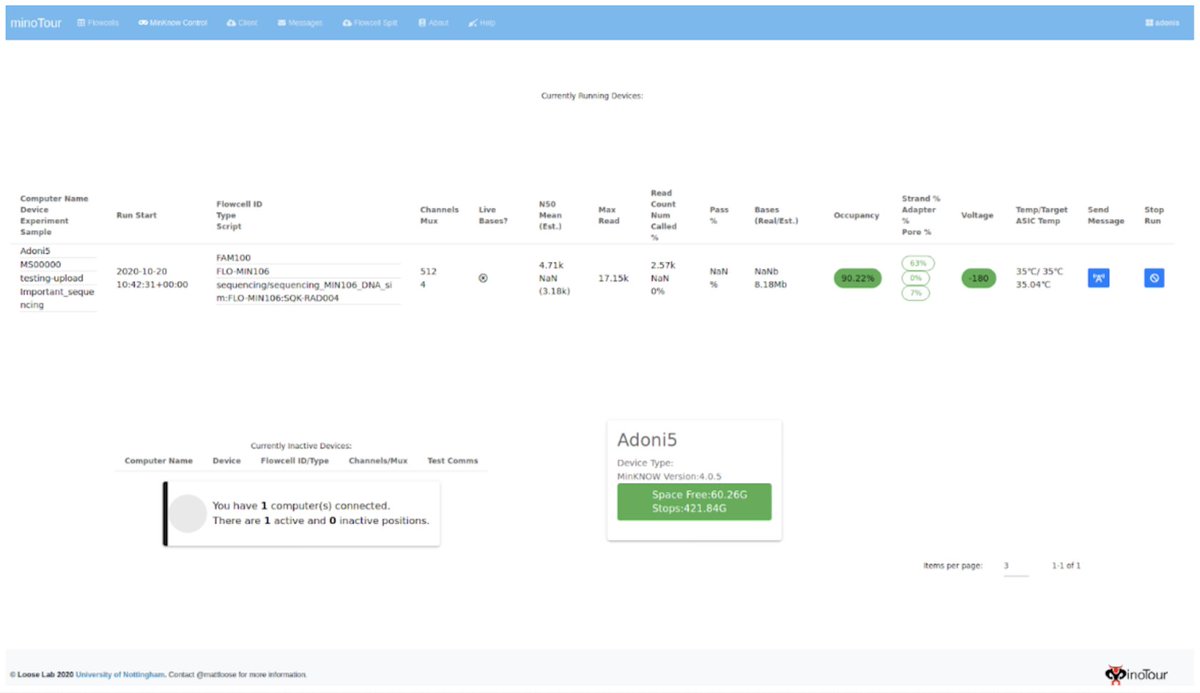



Obviously we track fastq data as well and so you can look at read length distributions and flow cell activity - separated by barcode if appropriate. 


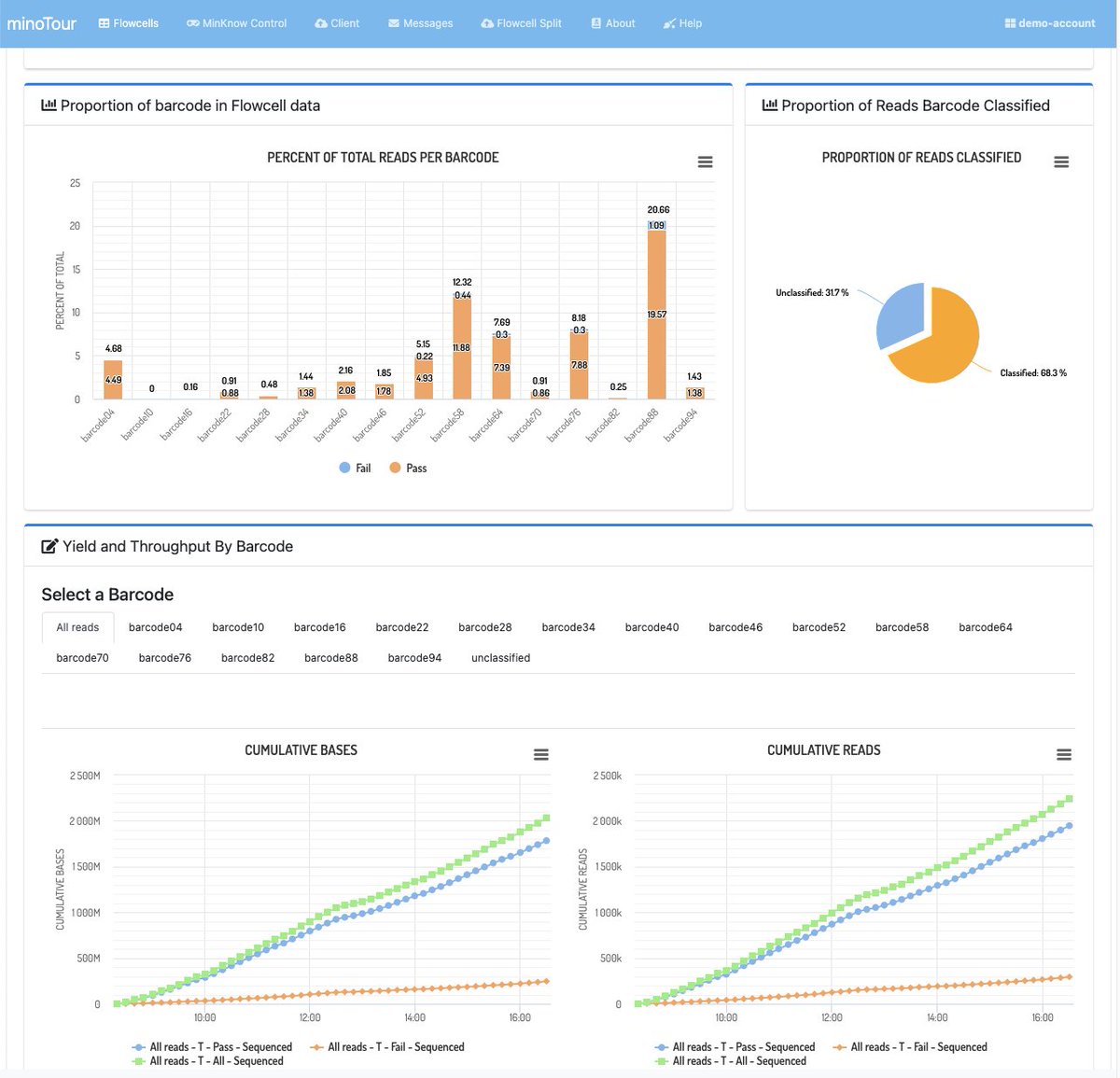
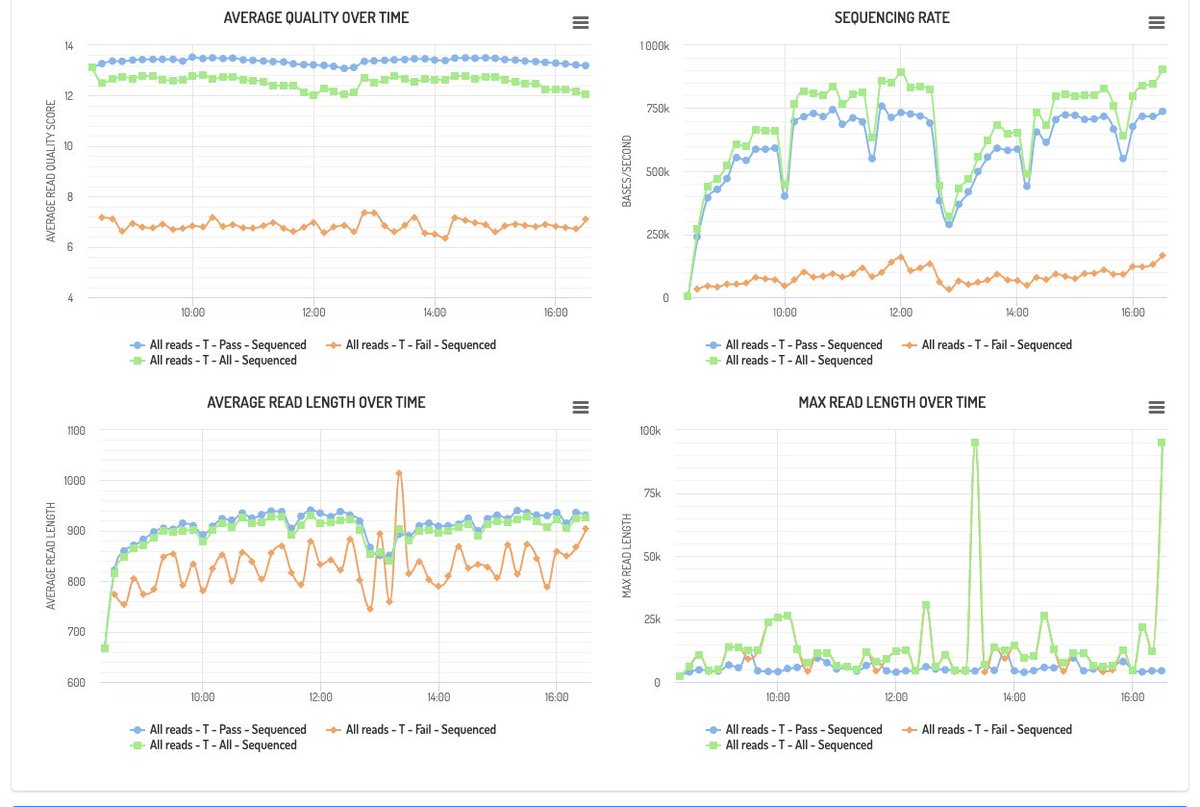

Data upload is handled by a python client which just runs in the background and provides metrics on whats happening. The client is available on PyPi. 

We can couple with our other tools, such as readfish (nature.com/articles/s4158…), to enable us to monitor enrichment in real time and visualise on and off target reads. 

Long term @nanopore users will have seen minoTour before - why so long for us getting to this point? Well - everytime we thought it worked throughput and yield would go up. Plus installation was not trivial.
MinoTour is now available through Docker and uses well established frameworks such as DJANGO, Celery, Redis etc (thanks @py5gol ).
When the pandemic hit, @Rorymatics started implementing the @NetworkArtic protocols into minoTour but those 400 bp reads come in fast! Re-engineering allowed us to call consensus genomes, assign VoC/VuIs and more in real time as sequencing was on going. 





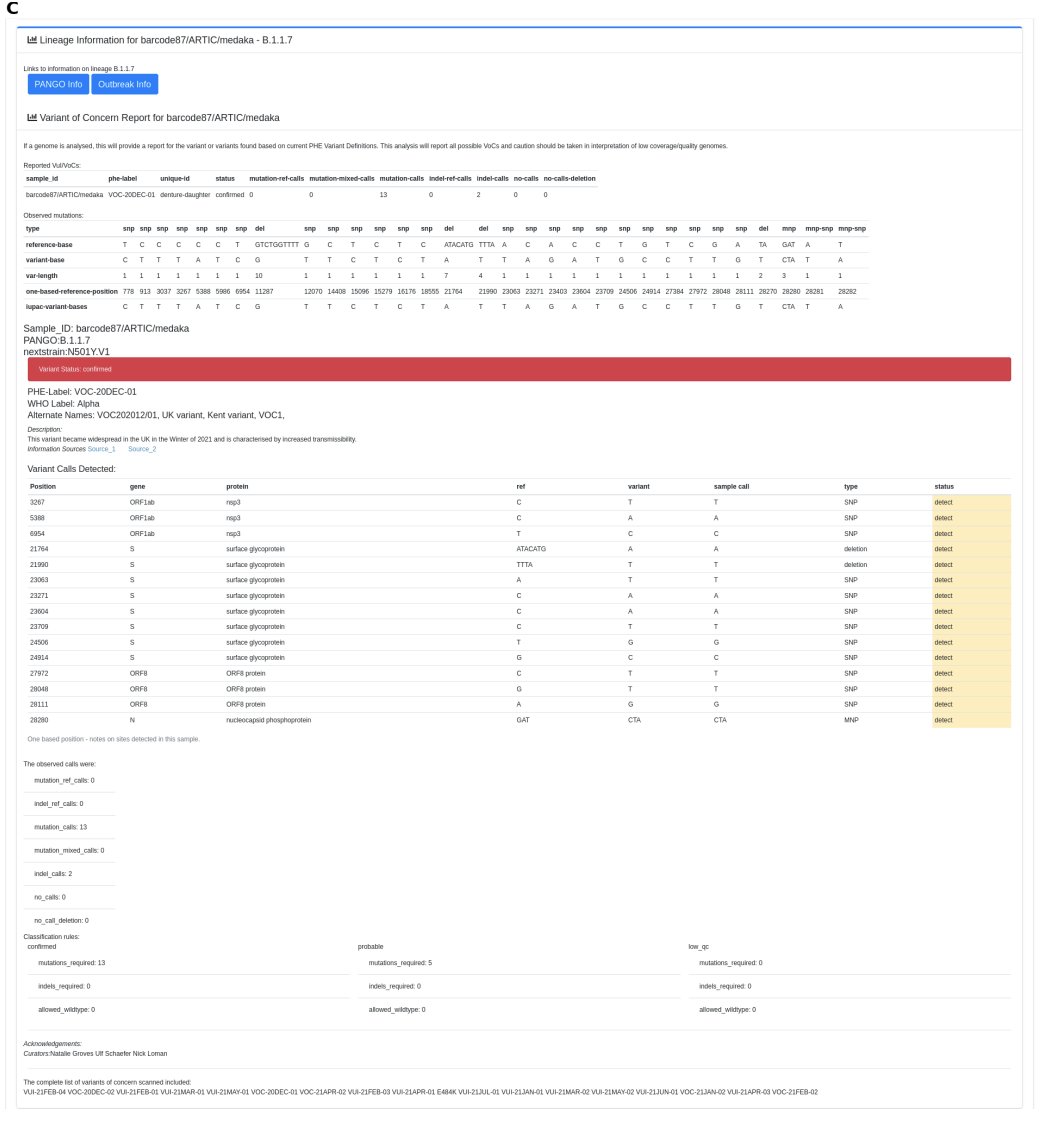

MinoTour predicts all sorts of information about completion rates for genomes and automatically fires consensus generation at the optimal time - see the paper for information. You can even receive a DM if a VoC/VuI is reported (though this is less helpful since Delta). 
Plus we can link out to outbreak.info and cov-lineages.org - minoTour can run multiple different artic compatible strategies at any time and users can upload there own schemas.
There's lots more in the papers - a demo server is available at http://137.44.59.170 with login details at github.com/LooseLab/minot… but at this time we don't run a single central hub for the community - users will need to set up their own (see readme).
Thanks to @Rorymatics , @py5gol , @DeepSeqNotts , @BioTeri , @alexomics and all others who have helped us with this. Also thanks to @NetworkArtic for providing fantastic resources.
• • •
Missing some Tweet in this thread? You can try to
force a refresh







