
Today we publish a paper in @ScienceMagazine that expands nanopore readings to the proteome:
a nanopore-based scanner to read off PROTEINS at the single-molecule level! 🤩
Awesome experiments by postdoc Henry Brinkerhoff of our #CDlab, with MD simulations of @aksimentievLab
1/x
a nanopore-based scanner to read off PROTEINS at the single-molecule level! 🤩
Awesome experiments by postdoc Henry Brinkerhoff of our #CDlab, with MD simulations of @aksimentievLab
1/x
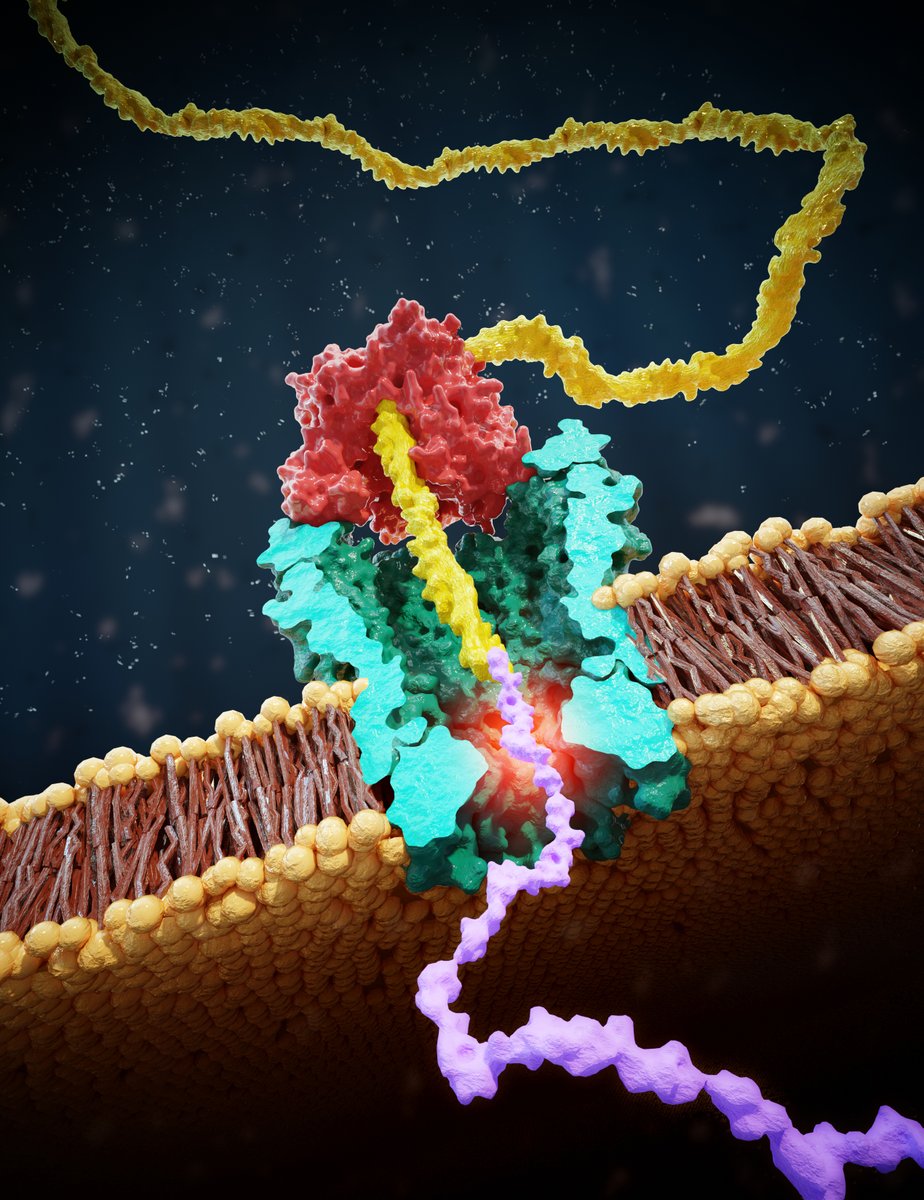
@ScienceMagazine @aksimentievLab Here’s the link to this paper in @ScienceMagazine entitled “Multiple re-reads of single proteins at single-amino-acid resolution using nanopores”: science.org/doi/10.1126/sc…
2/x
2/x

@ScienceMagazine @aksimentievLab Principle reminds of nanopore DNA sequencing: we draw a peptide through a nanopore with a helicase walking on a lead DNA strand, and then read off ion current step signals as amino acids are blocking the pore.
3/x
3/x

@ScienceMagazine @aksimentievLab This allows us to discriminate even single amino-acid substitutions in single reads of a peptide, with already very good fidelity (87%) in these first experiments
4/x
4/x

MD simulations by our collaborators @aksimentievLab show the (sometimes counterintuitive) ion current signals result from size exclusion as well as pore binding by amino acids.
5/x
5/x

@aksimentievLab Strikingly, we can RE-read one and the same molecule hundreds of times, which drives the read accuracy to 100% with an error rate for single amino-acid variant identification of < 1 in 10^6 on 1 peptide molecule – alleviating a major problem in nanopore sequencing!
6/x
6/x

Here’s for example a raw data. The second read is the one with a peptide, and it shows ~5 re-reads!
Isn’t that totally awesome…? #Iloveit
7/x
Isn’t that totally awesome…? #Iloveit
7/x
Anyway, I’m pretty excited about all this. Much much more to do of course, to develop these first proof-of-principle data on protein identification into a full de novo nanopore single-molecule protein sequencer, but a very exciting development imho.
science.org/doi/10.1126/sc…
8/8
science.org/doi/10.1126/sc…
8/8

• • •
Missing some Tweet in this thread? You can try to
force a refresh



