
As promised, a thread on our paper on bats and coronaviruses just published in @NatureRevMicro doi.org/10.1038/s41579…
A team effort from the BatOneHealth group: @MRuizAravena @clifmckee @AmandineGamble @lunn_tamika @yeoyaoyu @elinor_jax @cl_faust @dev_n_jones @maureenkkessler @caylee_falvo @danewingcrowley 

@nitanother @caraebrook @AguilarVirology @ali_bat @BugsWormsNBats @TSchountz @ColinP3 @EmilyGurley3 @jlloydsmith @PeterPjh18 @rainamontana
While planning new research on coronaviruses in #bats our team found information dispersed in hundreds of papers, so we decided to compile and review the literature so it would be useful for the global community of researchers
Our collective experts, including ecologists, immunologists, virologists, epidemiologists, modelers, and other disciplines, critically reviewed the knowledge on the ecology, evolution, and spillover of bat coronaviruses, including virus-cell interactions to #pandemic emergence
We reviewed over 300 articles, almost all published since the emergence of SARS-CoV-1 in 2002-2003 when Rhinolophus bats were identified as the main hosts of SARS-related viruses. Prior to that point, no groups were studying coronaviruses in bats.
Yet research on this subject has accelerated, so now there are dozens of bat #coronavirus papers published each year. 

More than 5,000 coronavirus sequences have now been detected in bats mgc.ac.cn/DBatVir/.
543 out of 1,435 known bat species have been sampled for CoVs in the wild and 238 (16%) have been found carrying one or more CoVs in the wild, representing 14 of the 21 bat families.
543 out of 1,435 known bat species have been sampled for CoVs in the wild and 238 (16%) have been found carrying one or more CoVs in the wild, representing 14 of the 21 bat families.

Now it is well established that bats are the main ancestral host group of alpha- and betacoronaviruses, and 5 of the 7 coronaviruses capable of human-to-human transmission can be traced back to lineages of viruses circulating in bats.
Bats host CoVs on every continent they inhabit. While sampling of bat species for CoVs largely reflects patterns of bat diversity (tropical hotspots), the distribution of positive hosts differs slightly, with lower density of host species in S America and more in SE Asia. 

Biological differences in bat-CoV interactions with certain bat families (Rhinolophidae) might explain some of this difference, but small sample sizes in some species in the Americas and more intensive sampling in China and SE Asia likely contribute as well.
The history of spillovers of bat CoVs into humans and other species is long.
The diversity of bats, their long evolutionary history (>60M yrs), and their gregarious behavior may facilitate a high diversity of CoVs, and opportunities to evolve by jumping among host species.
The diversity of bats, their long evolutionary history (>60M yrs), and their gregarious behavior may facilitate a high diversity of CoVs, and opportunities to evolve by jumping among host species.

But... For a successful spillover to occur, coronaviruses must percolate through a series of barriers, detailed in doi.org/10.1038/nrmicr….
Let’s break it down:
Let’s break it down:
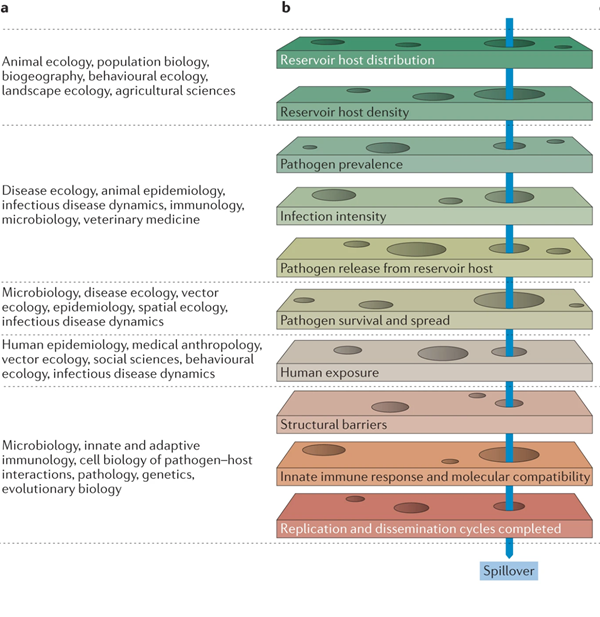
Spillover requires that bat populations be infected with and shedding of CoVs.
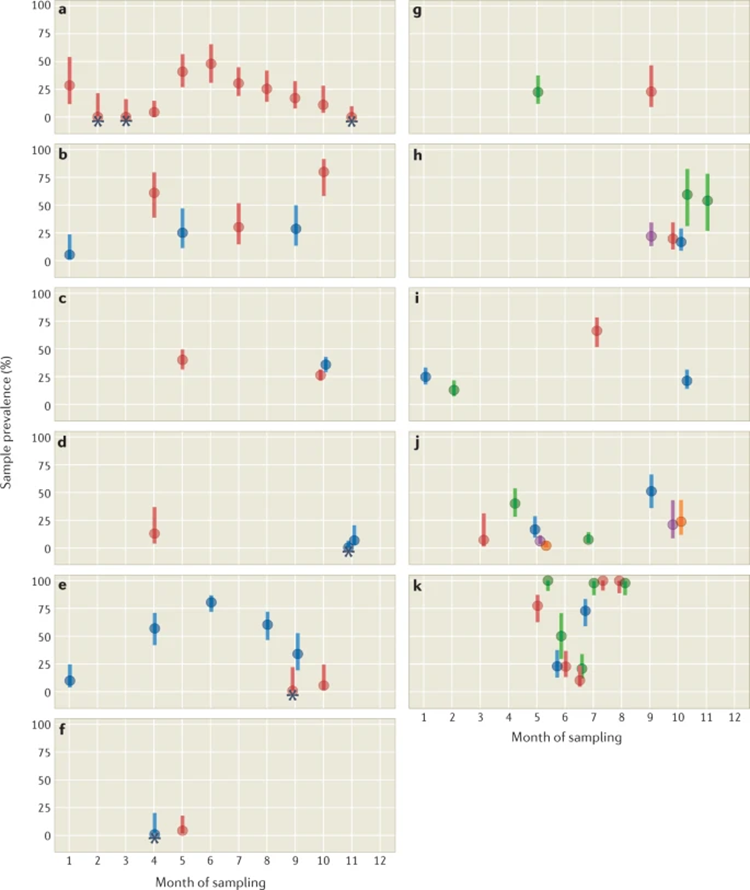
Few studies have surveyed bats over space and time to describe infection and shedding. In fact, only one study was truly longitudinal, but it still pooled samples over space
doi.org/10.1186/s12985…
Few studies have surveyed bats over space and time to describe infection and shedding. In fact, only one study was truly longitudinal, but it still pooled samples over space
doi.org/10.1186/s12985…

We found that 14 spatiotemporal studies had been done and they showed variable prevalence and shedding over time.
Often no CoVs are found, sometimes there is a pulse in CoV shedding. This is consistent with other bat virus systems.
Often no CoVs are found, sometimes there is a pulse in CoV shedding. This is consistent with other bat virus systems.
However, no studies combined the necessary data on bat ecology and health and environmental variables to understand the causes of CoV shedding pulses.
Without these data, we are "flying blind" with regard to understanding when and where spillover of bat CoVs might occur.
Without these data, we are "flying blind" with regard to understanding when and where spillover of bat CoVs might occur.
The 15+ years of research on CoVs in bats have given us valuable knowledge about the diversity and evolution of coronaviruses, the molecular machinery they use to enter host cells, and the factors that constrain their host ranges.
Yet much of this understanding is based solely on sequence data. We know little about how bats deal with infection in experimental settings. We found that only 6 studies have performed experimental infections of live bats. 

In particular, we have limited information about virus dynamics *within* bats, which may differ considerably from what happens in humans.
What receptors and proteases do bat CoVs use in bats?
Where are they expressed?
How do their immune responses control infection (or not)?
What receptors and proteases do bat CoVs use in bats?
Where are they expressed?
How do their immune responses control infection (or not)?
Ideally, we would have papers like this one for all bat hosts, potential bridge hosts, and relevant receptors nature.com/articles/s4159…
This is partly due to the difficulty of maintaining captive colonies of bats, but also a lack of isolated cultures of coronaviruses from bats necessary for experiments.
Isolating coronaviruses from wild bats appears to be exceedingly difficult!
Isolating coronaviruses from wild bats appears to be exceedingly difficult!
Out of 187 studies that examined viruses in samples from wild bats, only 42 (22%) actually attempted virus isolation in one or more cell cultures, and only 5 were successful, each producing a single isolate.
To our knowledge, only 5 coronaviruses have been isolated from bats.
To our knowledge, only 5 coronaviruses have been isolated from bats.
These include 1 related to MERS-CoV, 3 related to SARS-CoV-1, and 1 related to SARS-CoV-2.
The remaining diversity of CoVs in bats is based solely on sequencing and no physical isolate exists, including the famous RaTG13.
The remaining diversity of CoVs in bats is based solely on sequencing and no physical isolate exists, including the famous RaTG13.
Importantly, we know precious little about where and how CoVs are jumping from bats into humans and other animals.
Direct contact with bats (hunting/trade, guano mining, cave tourism) may be important, but few studies have examined whether such exposures result in spillover.
Direct contact with bats (hunting/trade, guano mining, cave tourism) may be important, but few studies have examined whether such exposures result in spillover.

Where outbreaks have occurred, with SARS and MERS, CoV spillover from bats was facilitated by a bridge host (civets, camels) that contact humans more frequently.
Other close interactions with domestic and captive animals could lead to spillovers without us knowing.
Other close interactions with domestic and captive animals could lead to spillovers without us knowing.
A big issue is that our surveillance systems are inadequate to detect uncommon spillover events. Rural settings where contact with bats or bridge hosts are higher may lack access to health care, so many subclinical infections and nascent person-to-person spread are undetected.
So what do we need to prevent CoV spillovers from bats from turning into the next pandemic?
Regarding ecology, we need more longitudinal studies, replicated over space, with collection of data on bat demographics, health, and environmental covariates to understand when, where, and why bats are shedding CoVs in the wild.
At the animal-human interface, increased surveillance of humans and animals in contact with bats and new tools like PhIP-Seq could provide consistent data on when and where spillovers are occurring and act as an early warning for epidemic emergence doi.org/10.1126/sciimm…
We need replicated studies of coronaviruses in live bats to understand the dynamics of coronavirus infection, evolution, and shedding in bat populations.
Development of computational and wet lab pipelines to translate sequence information from the field into assessments of a virus’s ability to infect human hosts could help to prioritize surveillance efforts doi.org/10.1038/s41564…
And lastly, the health of bats, the environment, and humans are all linked. We should proactively preserve and restore habitat and food resources for bats that could reduce the need for bats to use human-modified landscapes doi.org/10.1016/S2542-… 

@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh



