Up in a few minutes, Clive G Brown @the_taybor with a plenary talk on @nanopore sequencing. We'll live tweet in this thread 1/n #nanoporeconf 

@The_Taybor CB: our goal is to enable the analysis of anything, by anyone, anywhere #nanoporeconf
CB: A reminder of how nanopore works #nanoporeconf
CB: with @nanopore, any fragment length can be sequenced and accuracy is not read length dependent. #nanoporeconf
CB: @nanopore CAN sequence short fragments (<200bp) and show good results - the data is the same. We are now introducing software configurations to enable Short Fragment Mode to support as short as 20bp. You can do an F-tonne of short fragments #nanoporeconf
CB: New short read mode default to be released into MinKNOW in new year. 250M reads @~200bp possible from a single PromethION flow cell, with native human DNA, including a proportion of perfect reads #nanoporeconf 
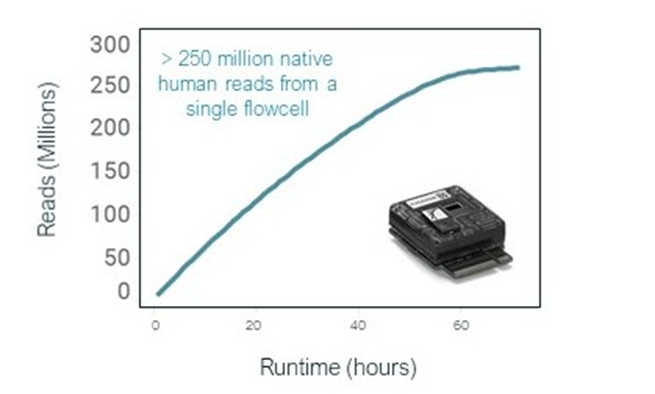
CB: We are now releasing a new kit – “kit 12”, which includes “Q20+” chemistry and is Duplex enabled. This enables >Q20 (>99%) modal raw read simplex accuracy with recently released R10.4 nanopore #nanoporeconf store.nanoporetech.com/sample-prep.ht… 

Apols @the_taybor, Clive's handle is @The__Taybor
CB: We have refined the movement of the motor - while it is a bit slower we get a significant shift in the baseline accuracy with Kit 12 and R10.4 #nanoporeconf
CB: diff types of accuracy; Duplex is enabled with kit12, enabling >99% modal raw read simplex accuracy with recently released R10.4 nanopore. Consensus is at Q50 on bacterial genomes, but coverage required is lower with kit12&R10.4 #nanoporeconf 

CB: We operate process of continuous improvement to algorithms, releasing features into research basecallers before moving to production. Eg Learnings from Bonito integrated into SUP basecaller in MinKNOW #nanoporeconf 

CB: Duplex – reading both forward and reverse strands immediately after one another – occurs at a natural baseline rate, but we can make amendments to increase this rate. #nanoporeconf 
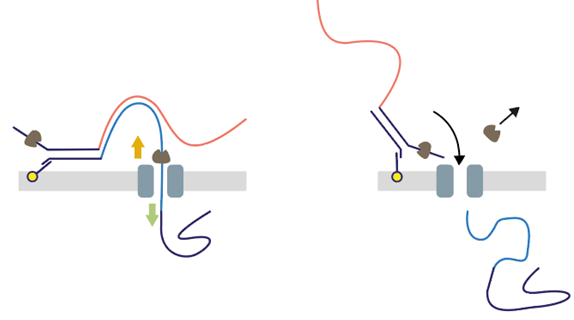
CB: Changes to kit 12 have increased Duplex rate, with internal data seeing up to 45% pairable data for accuracy around Q30 per single molecule
Pioneered by @ianholmes lab #nanoporeconf
store.nanoporetech.com/us/sample-prep…
Pioneered by @ianholmes lab #nanoporeconf
store.nanoporetech.com/us/sample-prep…

CB: The longest Duplex paired read to date has been 156 kb at ~Q30; there’s no inherent limit to duplex read length #nanoporeconf
CB: I wonder what ultra-long Duplex would do to assembly? Hard to believe this wouldn't have a significant effect, we hope to see more on this in the coming months #nanoporeconf
CB: Reminder: modification data is inherent in the nanopore signal, and so is available “for free”. We even showed data on this at AGBT 2012 #nanoporeconf
CB: Marcus Stoiber @stoibs11 will talk about this in more detail. However, in short we believe methylation data with R9.4.1 to be better than “gold-standard” bisulfite, and R10.4 is better than R9.4.1 #nanoporeconf 

CB: Making calling base modifications more accessible. With no more need to re-basecall to see modifications – new Remora tool runs alongside basecalling to annotate modifications (as a lightweight second pass). Remora is in Bonito now and Guppy in the new year #nanoporeconf 

CB: what's next? We have a hit list of modifications. This may be a bit vertebrate-centric so please speak up of you want others #nanoporeconf
CB: Accuracy improvements are not only in algorithms; there are lots of levers to pull and chemistry changes play a big part. Enzyme movement kinetics can be made more uniform, there are new versions in R&D pipeline #nanoporeconf 

CB: Innovations around the length of a nanopore drive accuracy even further. R10.4 better at homopolymers than R9.4.1. Next gen pores with even longer read heads are in research #nanoporeconf 

CB: The enzyme motor on DNA can also be influenced by changing temperature. Higher speeds may mean slight drop in accuracy, but more yield. Potential for on-flow cell control by users to select the data they want #nanoporeconf 

CB: Everything covered so far is in progress already. Time to look at more future-focused work #nanoporeconf
CB: There are basically two ways of putting DNA or RNA through pore: “Inny” or “Outy” (like a belly button). Product is currently in “Inny” mode but assessing enzymes for “Outy” suitability. Benefit to Outy in e.g. size selection #nanoporeconf 

CB: here is the original AGBT presentation from 2012 so you can compare where we are now to that vision. f1000research.com/slides/4-1206 #nanoporeconf
CB: More detail on Outy: DNA threads through pore and molecule can be sized. And then:
#nanoporeconf
#nanoporeconf

CB: Outy can potentially be used to re-read a single molecule many times. Pause at end of the strand, motor releases, DNA drops back, caught again and repeat. So you could "to-and-fro" a single molecule for very high accuracy consensus on that single molecule #nanoporeconf 

Oh by the way, the catflap reference from earlier:
https://twitter.com/odedrechavi/status/1463251239286812673?s=21#nanoporeconf
CB: Adaptive sampling, where we can hold on to the molecule, and then going up and down that molecule until you have the accuracy you need: Adaptive Accuracy #nanoporeconf 

CB: By combining Adaptive Sampling with Adaptive Accuracy we could illuminate rare single molecule mutations at ultra-high accuracy. Find your needle in a haystack, then confirm its significance #nanoporeconf 

CB: This could be the essence of liquid biopsy in a range of samples..blood..river..air droplets – fish through molecules not of interest, and find the key ones to give your answer #nanoporeconf
CB: No big hardware changes needed to enable this concept - and there are potential applications in labs and environment – Anything, Anyone, Anywhere #nanoporeconf
CB: Our sample-to-answer concept is built on our existing flow cell tech, with a new sample chamber added above pores. Raw sample added there, samples prepared, DNA electrophoretically moved to the pores beneath #nanoporeconf #skunkworx 

CB: Same concept opens up possibility for sensing other types of molecules, eg small molecules, metabolites. #nanoporeconf
CB: Introducing a new device. We introduced this concept introduced at LC 2019, and now PromethION 2 is available for preorder. Human WGS for less than $1000, with ~$10k starter pack. Falls between Grid and P24 #nanoporeconf nanoporetech.com/products/p2 

CB: P2 Solo is a modular sequencing unit which uses separate compute. Compatible with GridION, easier to place in automation robots #nanoporeconf 

CB: We have a proposed change to browser basecalling framework also. Use standard web browser for operation of basecalling, still exploit local GPU resource. #nanoporeconf 

CB: Bonito proof of concept now live for browser-based basecalling. Have a go! bonito.epi2me.io #nanoporeconf
CB: Browser based operation will be at the heart of the intended Mk1C replacement – MinION Mk1D. Custom tablet case with MinION integrated. Register your interest here register.nanoporetech.com/minion-mk1d #nanoporeconf 

CB: we are supporting/subsidising users who are sequencing critically endangered species – 22 draft genomes have been uploaded (open source) in a few months, we'd like to see more from scientists working with species in the field. org.one/oo #nanoporeconf
CB: we'd like to see people using eDNA to make a positive impact in biodiversity #nanoporeconf
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh










