Omicron. It not what you think it is.
Omicron is a name given by the WHO not from a scientific reason but mostly for journalist not calling it “The south African variant #2”.
But BA.2 is not (!!!!) BA.1 as I explained in the thread attached.
Omicron is a name given by the WHO not from a scientific reason but mostly for journalist not calling it “The south African variant #2”.
But BA.2 is not (!!!!) BA.1 as I explained in the thread attached.
https://twitter.com/shay_fleishon/status/1480933750041161731
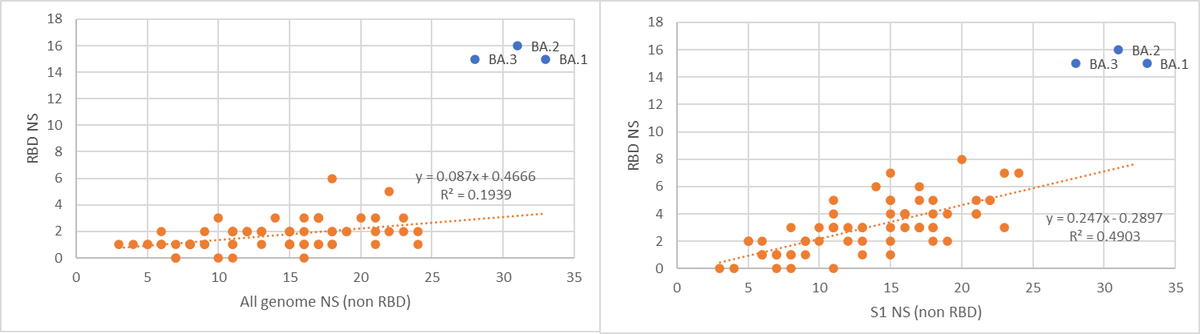
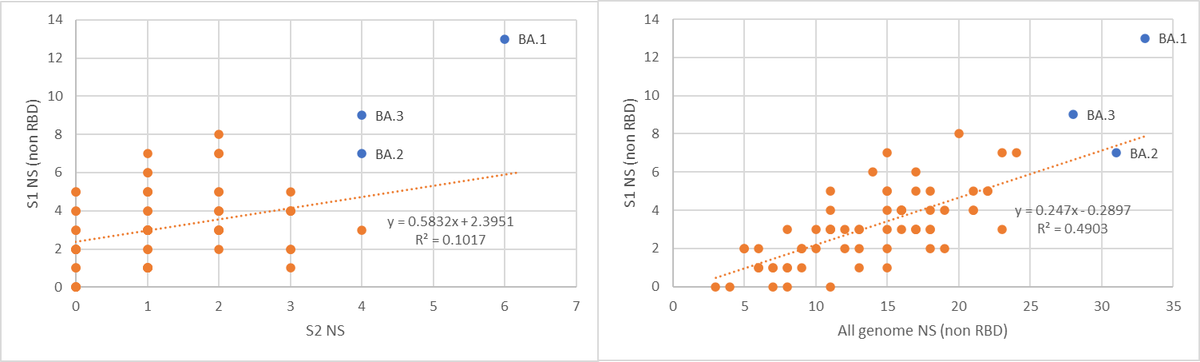
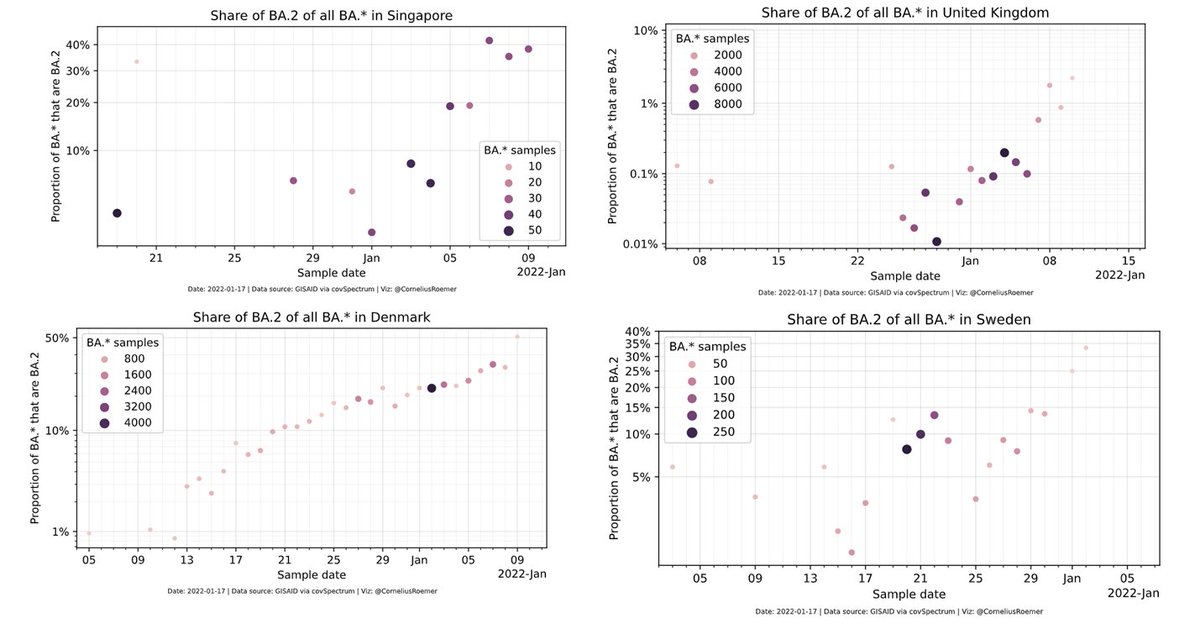
Is BA.2 has a higher Rt than BA.1? well the data as presented routinely by @JosetteSchoenma and by using the amazing app @CorneliusRoemer made shows it might even be more transmissible.
mybinder.org/v2/gh/corneliu…
mybinder.org/v2/gh/corneliu…
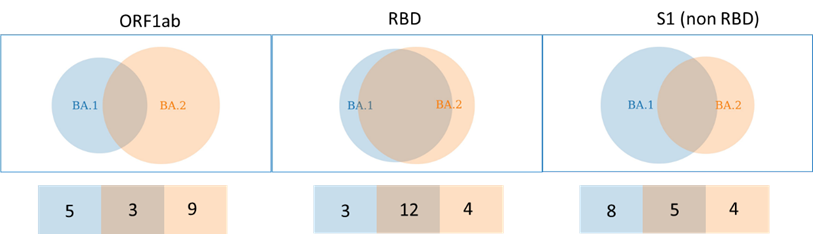
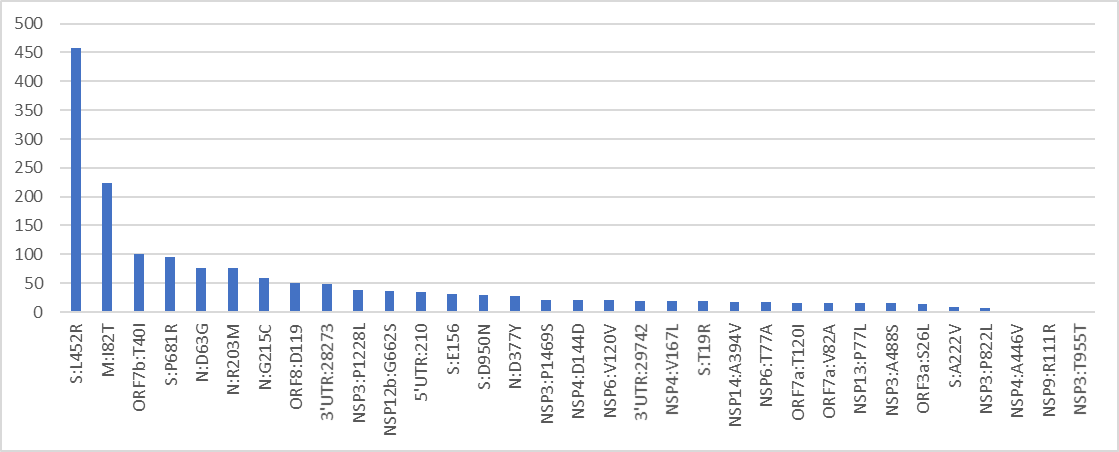
Can it evade our immune response? Well, look at it’s RBD, 16 NS SNP's, 12 are shared with BA.1.
No one will be surprised if it’ll be somewhere in the BA.1 level.
No one will be surprised if it’ll be somewhere in the BA.1 level.
Is it more severe? Well, I hope not.
But I also hope the Israeli national soccer team reach the finals at the world cup. In scientific work we should relate to hope as a bias factor.
We cannot know how different it's symptoms are until more data gathered. Just as was with BA.1
But I also hope the Israeli national soccer team reach the finals at the world cup. In scientific work we should relate to hope as a bias factor.
We cannot know how different it's symptoms are until more data gathered. Just as was with BA.1
So, I think the responsible thing to do is (*) to relate to BA.2 as a completely different variant, outcompeting BA.1. With all what it means.
(*) until more reliable clinical data gathered.
Oh and if someone in the WHO is here – The letter Pi is still waiting. Just saying.
(*) until more reliable clinical data gathered.
Oh and if someone in the WHO is here – The letter Pi is still waiting. Just saying.
• • •
Missing some Tweet in this thread? You can try to
force a refresh