
Interested in #phages, #metagenomics and #microbiome? New method out to bin #phages from shotgun metagenomics data by @jojohansen from our group. (1/12)
nature.com/articles/s4146…
nature.com/articles/s4146…
Discover and analyse #phages in any #Metagenomics shotgun dataset by:
1. Bin and identify high quality phage bins
2. Automatically defines viral populations across samples
3. Analysis of viral-host interactions within samples
1. Bin and identify high quality phage bins
2. Automatically defines viral populations across samples
3. Analysis of viral-host interactions within samples
Using two paired #metagenomics and #metaviromics datasets:
17-36% of HQ viruses in metavirome was found from metagenomics samples using PHAMB
46-89% of HQ viruses found by PHAMB in metagenome data was not found in the metavirome
17-36% of HQ viruses in metavirome was found from metagenomics samples using PHAMB
46-89% of HQ viruses found by PHAMB in metagenome data was not found in the metavirome
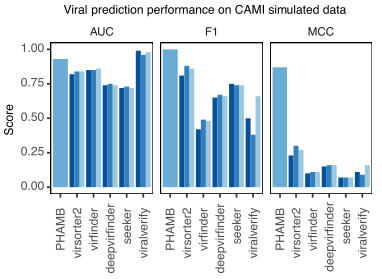
PHAMB has very high performance in estimating what is a true viral bin with MCC=0.87 compared to the next best (single contig) method viralVerify with MCC=0.39 
PHAMB builds upon VAMB, our #deeplearning binner for metagenomics data.
Therefore, PHAMB "learns" which viruses are the same across samples, ie. viral populations. This is really super cool as it allows analysis of viral populations across samples.
nature.com/articles/s4158…
Therefore, PHAMB "learns" which viruses are the same across samples, ie. viral populations. This is really super cool as it allows analysis of viral populations across samples.
nature.com/articles/s4158…
For instance known crAss-like bins were captured in five VAMB clusters that were phylogenetically consistent 

We used the method to analyse the #HMP2 #IBD cohort from where no high quality viral genomes were available until now. 130 individuals with 1317 samples.
The metavirome was highly stable and personal over time, and the viral signal clearly differentiated IBD dysbiosis.
The metavirome was highly stable and personal over time, and the viral signal clearly differentiated IBD dysbiosis.

Because we identify both bacterial (MAGs) and viral bins we can connect them. For instance, Faecalibacterium and Bacteroides MAGs were viral hotspots where ~99% of the MAGs was associated with a HQ viral bin. 

Furthermore, we could investigate genes carried by the phages and the dark-matter virome. Here we found more than 43k viral-like bins with confident CRISPR spacers in our dataset suggesting unsampled phage diversity in the gut microbiome.
Phamb is freely available at github.com/RasmussenLab/p…
Vamb is freely available at github.com/RasmussenLab/v…
Thanks to the collaborators:
Søren Sørensen and Dennis Sandris Nielsen from UCPH.
Shiraz Shah and the @COPSAC_com team for super cool paired metagenome and metavirome data.
Søren Sørensen and Dennis Sandris Nielsen from UCPH.
Shiraz Shah and the @COPSAC_com team for super cool paired metagenome and metavirome data.
And I tagged Joachim wrong again, it should be @JoJohanse
• • •
Missing some Tweet in this thread? You can try to
force a refresh



