How precise are noisy #morphogen gradients in #tissue #patterning?
It turns out that the positional information they convey to #cells in the #neuraltube is much more accurate than previously estimated!
Our new #devbio paper is out in @NatureComms: rdcu.be/cH8Fx
It turns out that the positional information they convey to #cells in the #neuraltube is much more accurate than previously estimated!
Our new #devbio paper is out in @NatureComms: rdcu.be/cH8Fx

A methodological subtlety has previously led to overestimated positional errors of the morphogen gradients in the mouse neural tube. We reanalysed the noisy gradients and found that the positional information they can convey to cells is actually orders of magnitude more precise. 
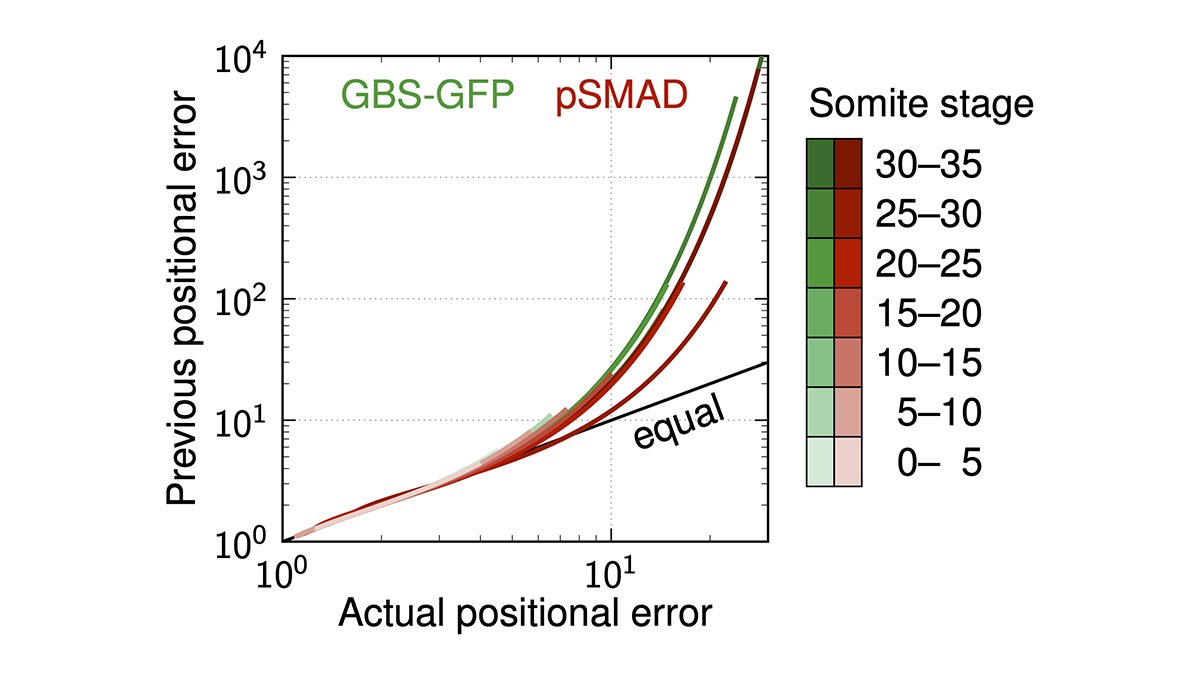
Also, morphogen gradients in differently sized tissues must not be pooled and rescaled to their mean length, or else an artificial positional error is added. 

If corrected for these effects, single morphogen gradients in the mouse neural tube are sufficiently accurate to explain the observed precision of progenitor domain boundaries in the center of the tissue. 

With 8-bit imaging, exponential gradients cannot be resolved over more than 5.5 times the gradients’ decay length (gray shade). Beyond that, only noise remains. To measure their shape over longer distances, higher imaging bit depth is needed. 

We developed a formula to estimate the positional error in those parts of the tissue that cannot easily be imaged from gradient variability close to the source.
Check out our tweetorial on this:
Check out our tweetorial on this:
https://twitter.com/DagmarIber/status/1385128539410870272

We also developed a novel simulation framework to estimate morphogen gradient variability from measured molecular noise levels.
We find that for reported noise levels, the expected positional error of gradients does not exceed the reported positional error of their readouts.
We find that for reported noise levels, the expected positional error of gradients does not exceed the reported positional error of their readouts.

In a related paper, we find that the #precision of morphogen gradients is strongly affected by #cell #size. Higher patterning precision is achieved with smaller cells:
Preprint: doi.org/10.1101/2022.0…
https://twitter.com/DagmarIber/status/1489479705292156933
Preprint: doi.org/10.1101/2022.0…

Our discovery will hopefully motivate further gradient measurements, and facilitate #tissue #engineering: Precise patterns can result from single morphogen gradients, which is great news for everyone seeking to synthetically recapitulate patterning processes. 

We have many colleagues to thank: Marius Almanstötter, @MarceloBoareto, @briscoejames, @fcasfer, Jose Dias, Johan Ericson, @gregorscience, @marco__meer, Thomas Horn, @AKicheva, Alice Kluzer, @F_Lampart, as well as the anonymous referees & the editors @NatureComms who supported us
• • •
Missing some Tweet in this thread? You can try to
force a refresh



