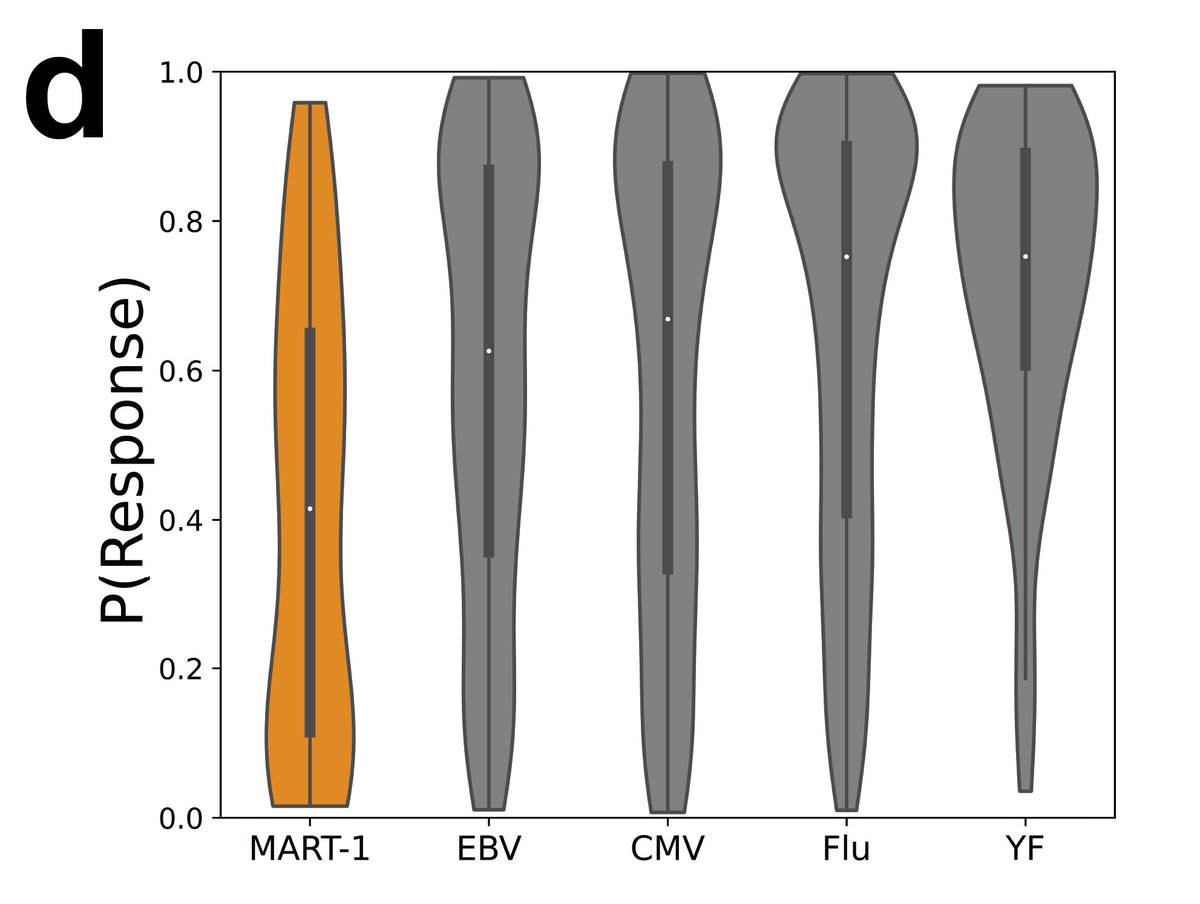
5 years ago, I proposed using #DeepLearning to predict response to immunotherapy in TCR-Seq.
🧬🧬🧬🧬
Last year, we published #DeepTCR. Today, I am proud to share its application in cancer immunology, now out in @ScienceAdvances #ScienceResearch
🔶🔷🔶🔷
science.org/doi/10.1126/sc…
🧬🧬🧬🧬
Last year, we published #DeepTCR. Today, I am proud to share its application in cancer immunology, now out in @ScienceAdvances #ScienceResearch
🔶🔷🔶🔷
science.org/doi/10.1126/sc…
This project started with a hypothesis that if a specific antigen-specific response was characteristic of responses to immunotherapy, then this should be reflected in the TCR repertoire/sequences.
After all, the TCR repertoire is ground truth for the antigen-specific response.
After all, the TCR repertoire is ground truth for the antigen-specific response.
First, we expand the #DeepTCR framework by allowing featurization of the TCR in context of the HLA background it is found within, overcoming the challenge in making direct comparisons of TCR repertoire in HLA mismatched individuals. 
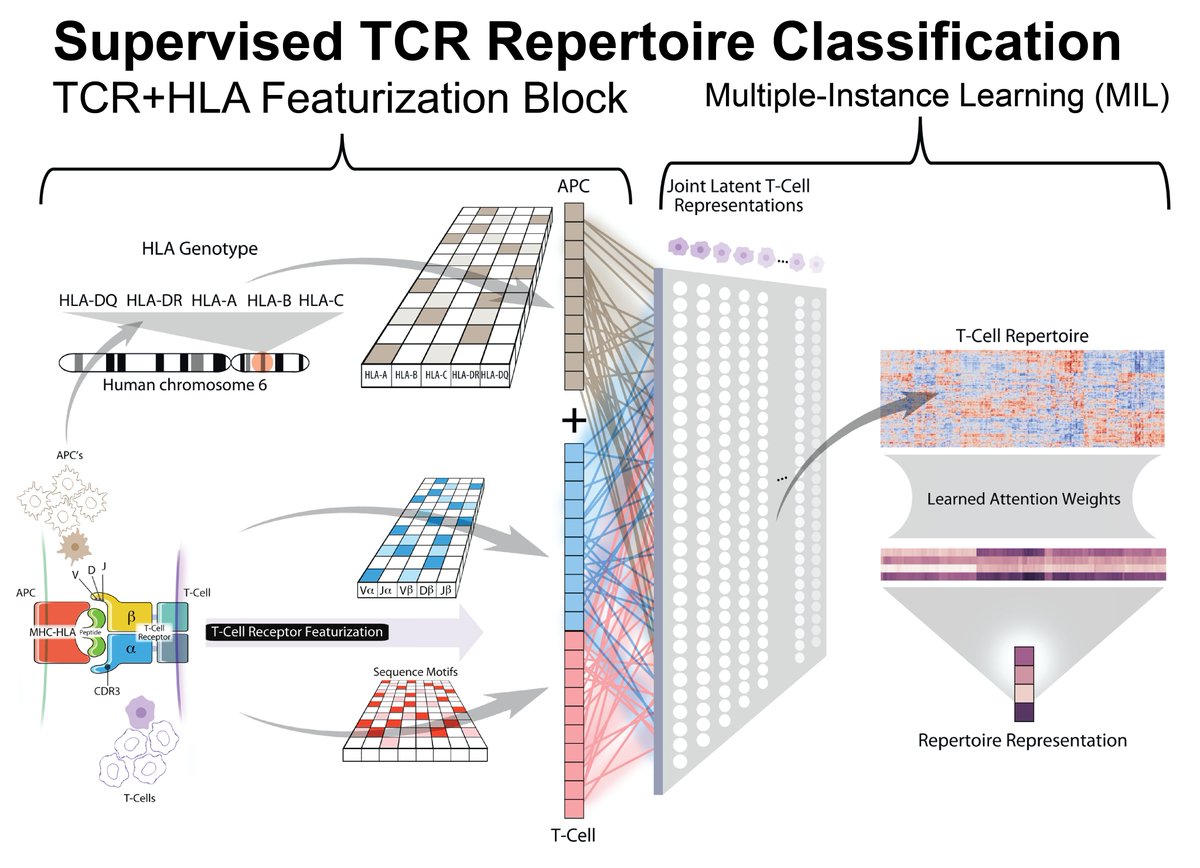
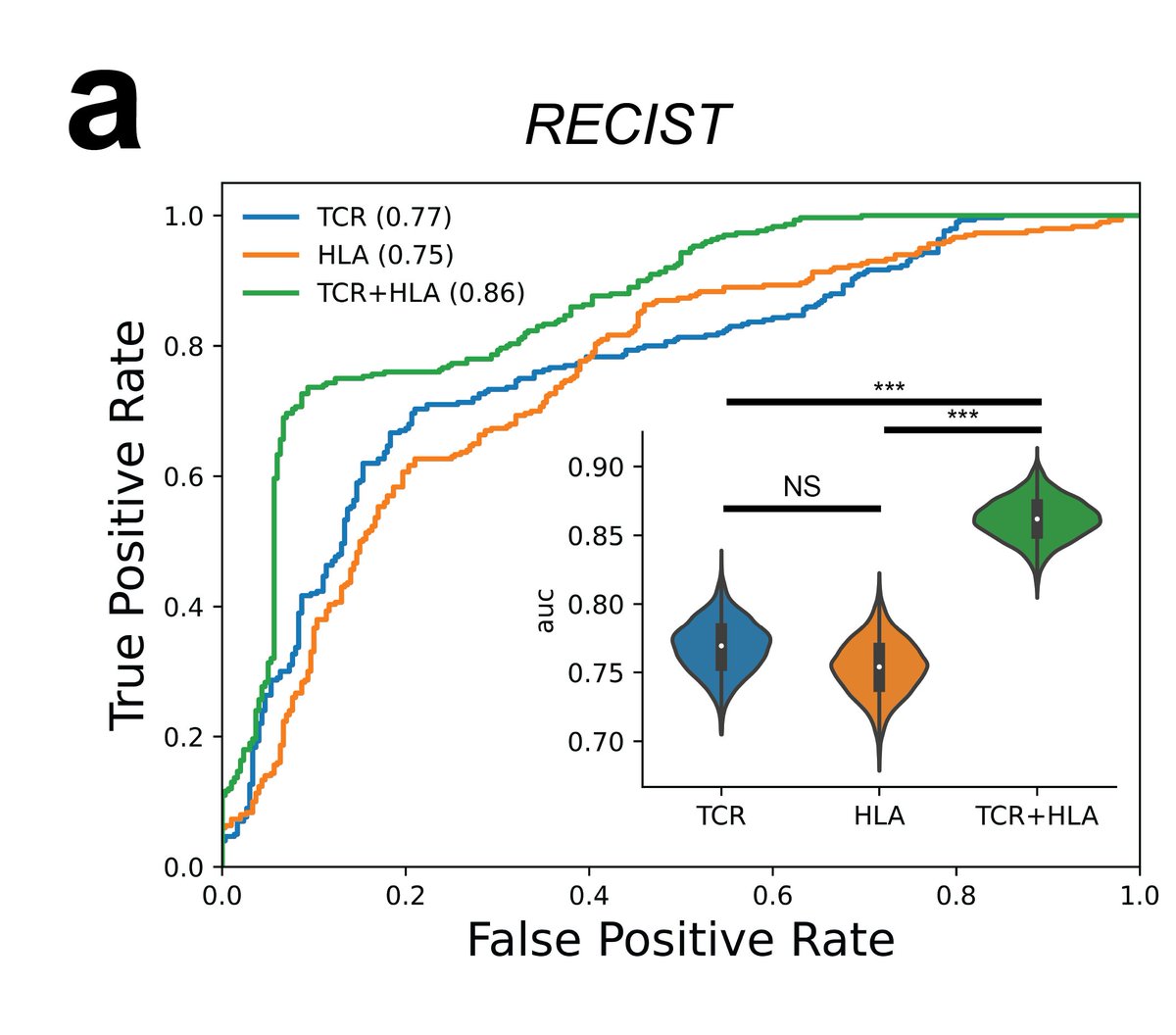
We show that when incorporating HLA and TCR-Seq information into one model, that we obtain a significantly higher performance to predict response to immunotherapy in #CheckMate038 from pre-treatment melanoma samples (as determined by the RECIST criteria). 
While we were excited to have demonstrated the first predictive model of response to immunotherapy in cancer from TCR-Seq sequence information, we were more intrigued as to figuring out the biological meaning of this predictive signature... 🤔
We first use the #DeepTCR variational autoencoder, a completely unsupervised method of TCR representation, + UMAP to visualize the distribution of the predictive TCRs in responders/non-responders, and noticed clearly shared concepts within these cohorts. 
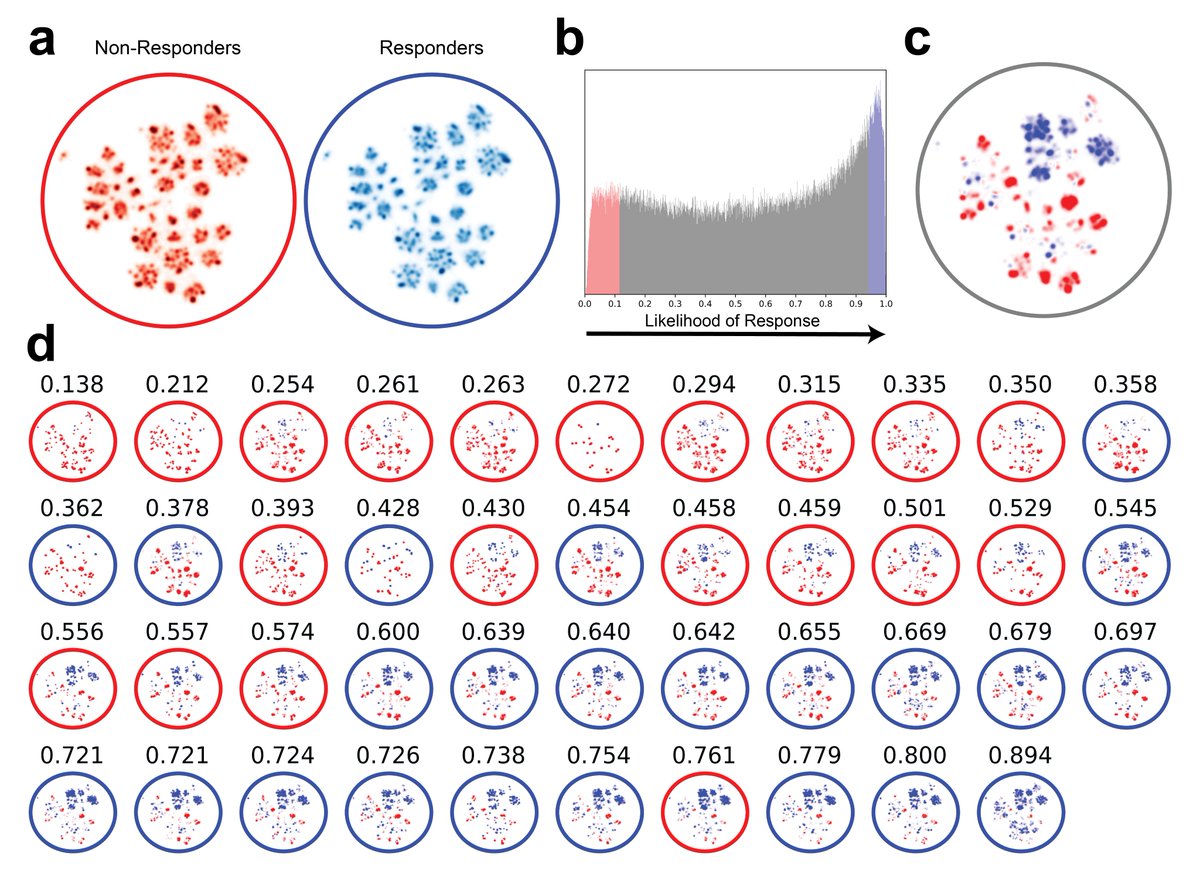
Notably, when looking at the TCR repertoire on treatment, we found the same strength of the predictive signature weeks into treatment. 

This finding left us with the inevitable question of "what is the antigen and/or antigens that this predictive signature corresponds with?"
To answer this question, we used a dataset from recently published paired single-cell TCR-seq with known antigens in melanoma (provided by Drs. Giacomo Oliveira and Catherine Wu at @DanaFarber @harvardmed). nature.com/articles/s4158…
When assigning P(response) to all TCRs found in their dataset from our predictive model, we noted that the predictive signature of response was surprisingly associated with the viral TCRs and the predictive signature of non-response was associated with tumor-specific TCRs. 



Further, when looking at the dynamics of these predictive TCRs during therapy, we noted a higher turnover of tumor-specific TCRs in non-responders at the clonotype and patient level. 
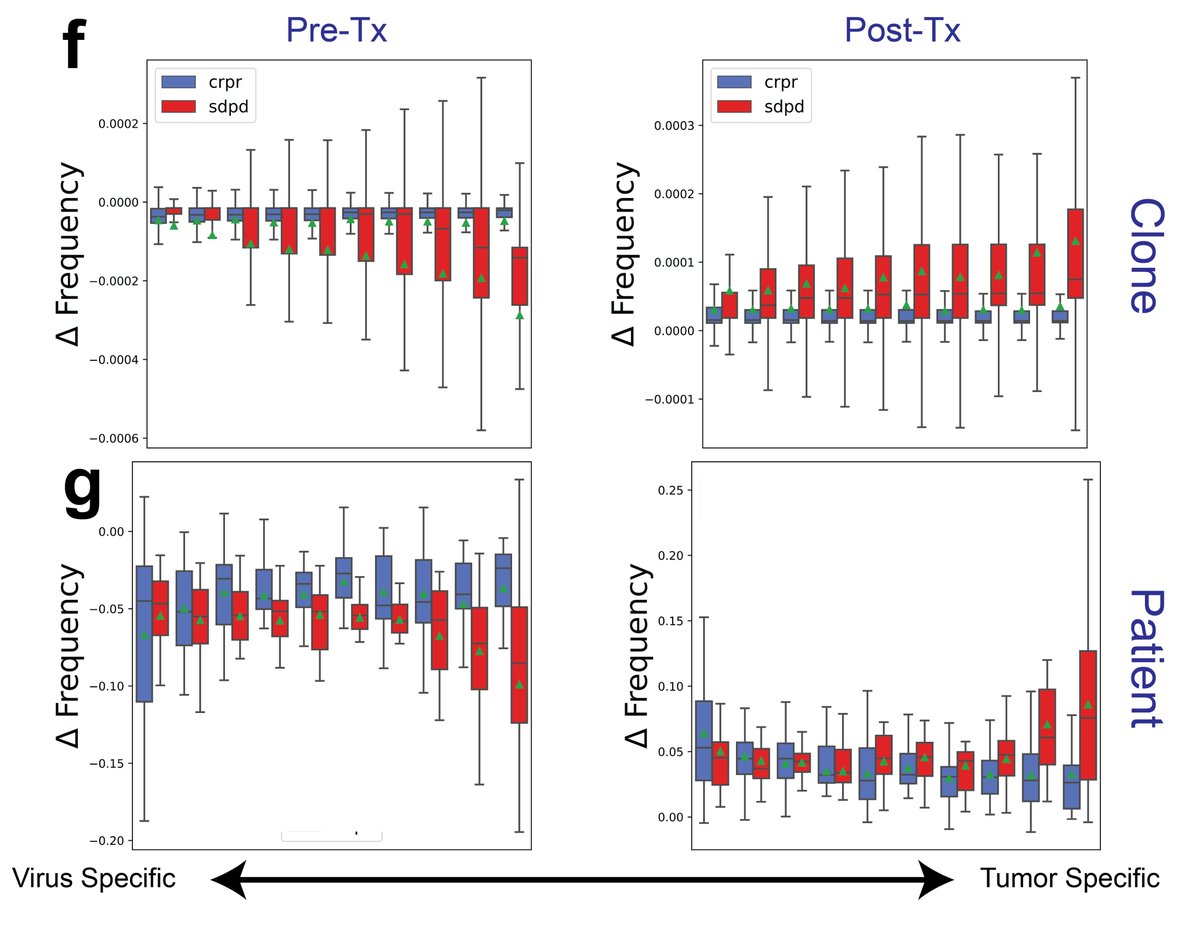
Taken together, we propose a model of the antigenic response and its dynamics under immunotherapy in responders vs. non-responders of immunotherapy, where non-responders exhibit an accumulation and higher turnover of tumor-specific clones, likely due to their dysfunctional state. 
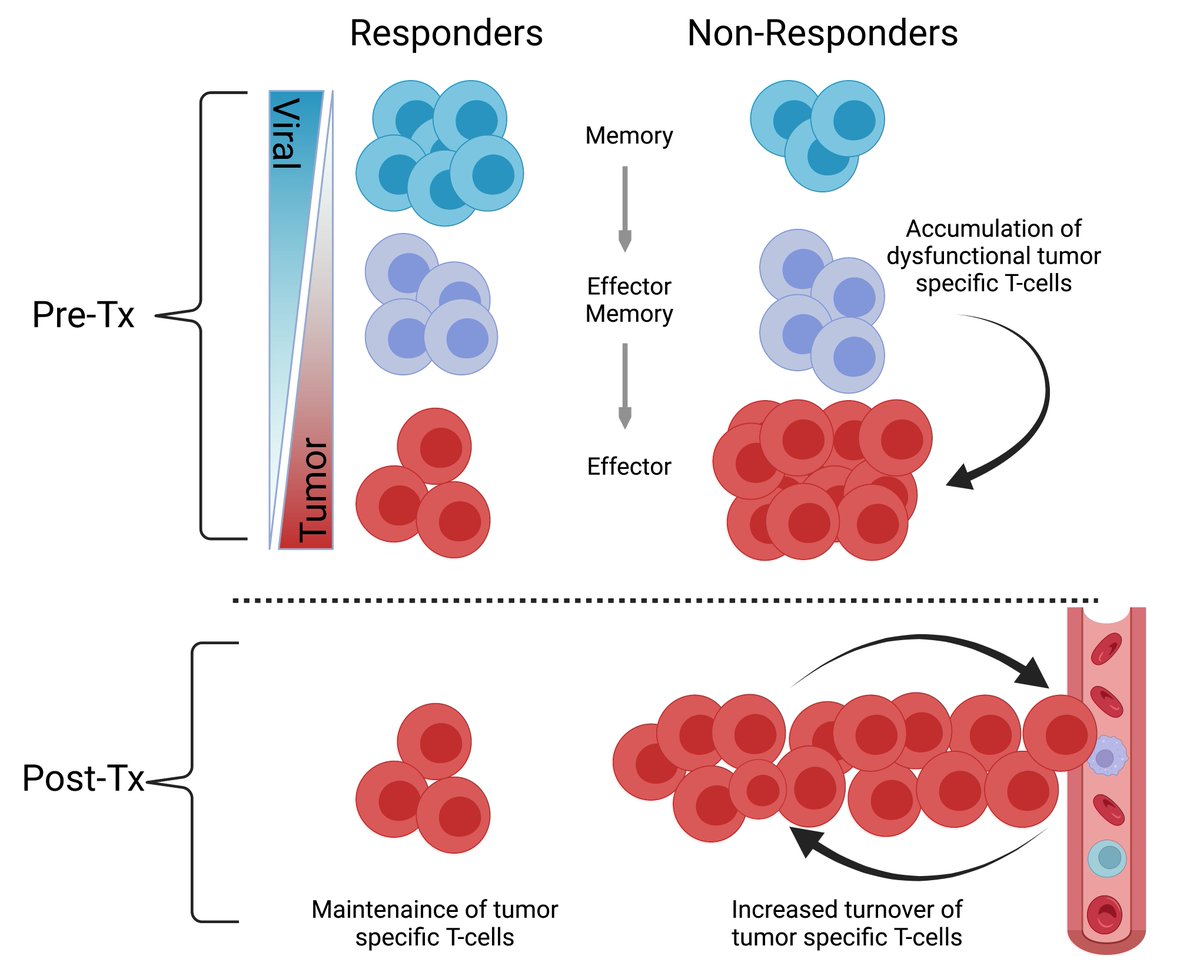
While this proposed model is merely our interpretation of the data, by no means is it a proven model. We would love to hear alternative interpretations of the data and resulting analyses!
While the findings presented are consequential for how we understand response to immunotherapy, I find this work to be more valuable as a demonstration of a systematic approach to dissect and understand immune repertoires with predictive models.
In addition, to see a brief oral presentation I gave at the @sitcancer Annual Conference in 2021 on this work, check out the page below!
#SITC2021
johnwilliamsidhom.com/talk/sitc_2021/
#SITC2021
johnwilliamsidhom.com/talk/sitc_2021/
This work wouldn't be possible w/o the mentors and collaborators at @HopkinsMedicine @HopkinsMDPhD @DanaFarber @harvardmed @bmsnews @ScienceAtBMS @IcahnMountSinai including @dpardol1 , @alexander_baras , Catherine Wu, Giacomo Oliveira, Megan Wind-Rotolo, and Petra Ross-MacDonald.
For reference, see below for the original #DeepTCR manuscript 👇
https://twitter.com/John_Will_I_Am/status/1370025771696201733?s=20&t=iuYGGN0L9hRo9uE7z3Vy_A
... as well as another manuscript we published last year using #DeepTCR to predict clinical outcomes in #COVID19 👇
https://twitter.com/John_Will_I_Am/status/1414622230600892419?s=20&t=k-AhMitTbafb7Oy28xqVpg
.. and if you are curious, check out the story of how #DeepTCR came to be including its initial inspiration and long, difficult path to make it a reality.
I could not be happier to add another chapter to its story today.
johnwilliamsidhom.com/projects/deept…
I could not be happier to add another chapter to its story today.
johnwilliamsidhom.com/projects/deept…
Finally, the timing of this application to #DeepTCR to immuno-oncology could not be more meaningful as I find myself in the midst of applying to heme/onc fellowship as well as on the inpatient liquid malignancy service at @TischCancer @IcahnMountSinai @MSH_MedChiefs
• • •
Missing some Tweet in this thread? You can try to
force a refresh