BREAKING🔔 The 14th paper from G2P-Japan🇯🇵 is out at Cell Host & Microbe @cellhostmicrobe. #Omicron BA.2.75 (aka #Centaurus) seems to be more contagious than BA.5 and more pathogenic than the original BA.2. Please RT. 1/16
cell.com/cell-host-micr…
cell.com/cell-host-micr…
BA.2.75, emerged in India🇮🇳, is classified as VOC-LUM (variant of concern linage under monitoring) by WHO on July 7. Here we elucidated the transmissibility, sensitivity to antiviral immunity & drugs, and intrinsic pathogenicity of BA.2.75. 2/16
who.int/activities/tra…
who.int/activities/tra…
BA.2.75 is a descendant of BA.2. However, BA.2.75 is phylogenetically different from BA.5, the currently predominant BA.2 descendant. BA.2.75 spike bears nine substitutions, but only a substitution is shared with BA.5 spike. 3/16 

First, our mathematical/statistical analysis revealed that the effective reproduction number ("Re" in the figure) of BA.2.75 is greater than that of BA.5. 4/16 

Second, we performed neutralization assay. we showed that:
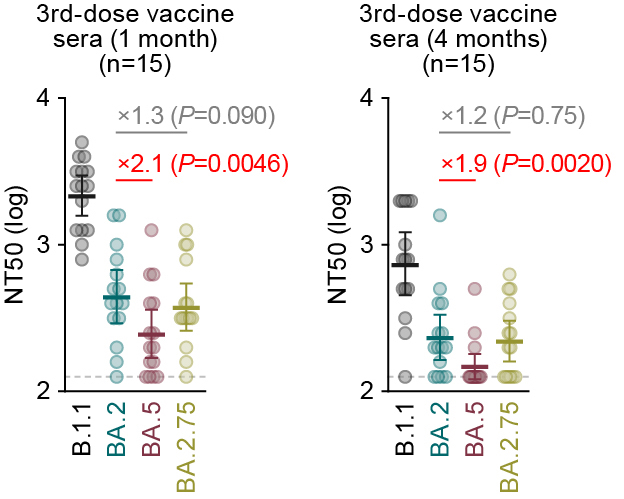
1⃣ Resistance to 3rd-dose vaccine sera: BA.2.75 = BA.2. 5/16
1⃣ Resistance to 3rd-dose vaccine sera: BA.2.75 = BA.2. 5/16
6⃣ Resistance to BA.2.75-infected hamster sera: BA.2.75 << BA.5 (6.4-fold!), suggesting that BA.2.75 is immunogenetically different from BA.5. 10/16 

7⃣ We further evaluated the efficacy of 10 therapeutic monoclonal antibodies against BA.2.75. FYI, please see the following thread↓ 11/16

https://twitter.com/SystemsVirology/status/1548069972294807552?s=20&t=AqA0QIOi5q4nHbiynM2apQ

Antiviral activity of three drugs, Remdesivir, Molnupiravir (EIDD-1931) and Paxlovid (Nirmatrelvir) against BA.2.75 was also evaluated. All 3 drugs were effective against BA.2.75. 12/16 

We solved the structure of BA.2.75 spike by cryo-electron microscopy and showed how BA.2.75 spike exhibits very high affinity to human ACE2 receptor. 13/16 



Using clinical isolates of BA.2, BA.2.75, BA.2 & Delta and a hamster model, we demonstrated that the intrinsic pathogenicity of BA.2.75 is comparable to that of BA.5 but is greater than that of the original BA.2. 15/16 

▶️ Our multiscale investigations suggest that BA.2.75 acquired virological properties independently of BA.5, and the potential risk of BA.2.75 to global health may be greater than that of BA.5. 16/16 

• • •
Missing some Tweet in this thread? You can try to
force a refresh