🎉CoVariants.org Update!🎉
23B (XBB.1.16) is now available on CoVariants! It's visible as part of Per Country & Per Variant plots, on the shared mutation page - and of course, has a page of its own.
1/10
#XBB116 #23B #Arcturus
📷
23B (XBB.1.16) is now available on CoVariants! It's visible as part of Per Country & Per Variant plots, on the shared mutation page - and of course, has a page of its own.
1/10
#XBB116 #23B #Arcturus
📷

This is in line with the recent addition of 23B (XBB.1.16) as a @nextstrain clade - read more on that below 👇🏻!
2/10
2/10
https://twitter.com/nextstrain/status/1650545883480567813
As I covered earlier, 23B (XBB.1.16) is descended from the recombinant 22F (XBB) variant, with some additional mutations. You can read more about how it evolved & acquired those mutations below 👇🏻.
3/10
3/10
https://twitter.com/firefoxx66/status/1648588097381101570
On the 23B page, you can read about some initial 23B (XBB.1.16) work on neutralizing titres, see a list of defining mutations, a plot of the variant growth, link to Aquaria protein viz, & see a list of other mutations present in the variant.
4/10
covariants.org/variants/23B.O…

4/10
covariants.org/variants/23B.O…


There's also a link to the 23B (XBB.1.16) @nextstrain
focal build: nextstrain.org/groups/neherla…
Here, we can zoom in on the clade & get a closer look at how 23B sequences are distributed around the world, & their diversity. 🌏🌍
5/10
#XBB116

focal build: nextstrain.org/groups/neherla…
Here, we can zoom in on the clade & get a closer look at how 23B sequences are distributed around the world, & their diversity. 🌏🌍
5/10
#XBB116
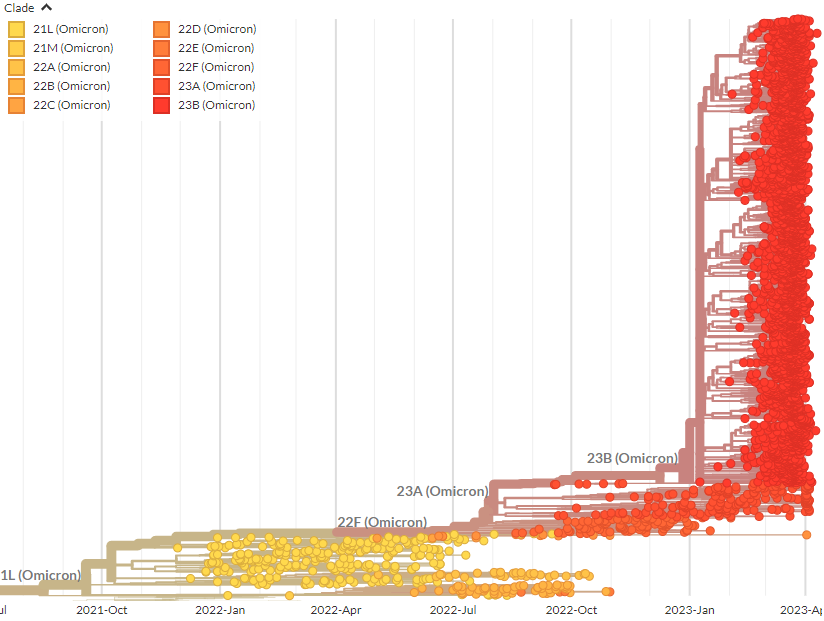

On the Per Country page, 23B (XBB.1.16) is already clearly visible in India & Singapore, & is starting to appear in smaller numbers in other countries, like Australia & the US. Remember, sequencing data is always a few weeks behind.
6/10
covariants.org/per-country
6/10
covariants.org/per-country

Finally, on the Shared Mutations page, you can see how 23B's / XBB.1.16's Spike mutations compare to other VoC, including its Omicron family, 22F (XBB) parent, and its 'sibling' 23A / XBB.1.5.
7/10
covariants.org/shared-mutatio…
7/10
covariants.org/shared-mutatio…

More information on 23B (XBB.1.16) & its spread will become available in due course.
As always, CoVariants.org is open-source & we welcome your PRs & suggestions to add more studies & information to variant pages! (Or any error-catching!)
github.com/hodcroftlab/co…
8/10
As always, CoVariants.org is open-source & we welcome your PRs & suggestions to add more studies & information to variant pages! (Or any error-catching!)
github.com/hodcroftlab/co…
8/10
As before, I've created a file for 23B (XBB.1.16) mapping all defining mutations (relative to ancestral), including nuc->AA. This is available on the CoVaritants Github!
Huge 🙏🏻to all who helped check it! (Esp @CorneliusRoemer @AngieSHinrichs)!
github.com/hodcroftlab/co…
9/10
Huge 🙏🏻to all who helped check it! (Esp @CorneliusRoemer @AngieSHinrichs)!
github.com/hodcroftlab/co…
9/10
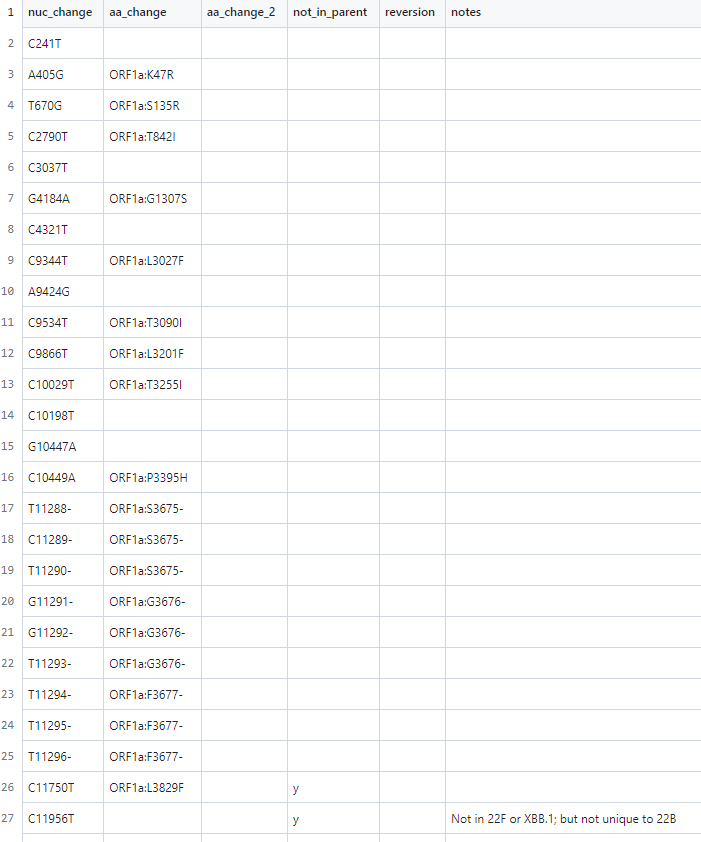
The eagle-eyed among you will have noticed that I've adjusted the 23A colour when adding 23B - the colours were probably always a bit too close together but this became even more obvious with 23B. I hope it doesn't cause any confusion! 🌈
10/10
10/10

• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter