Here's a short (?) thread on a recent #PCAWG paper focused on #cancer, long-reads, tumor mutational burden, and a very hard-to-pronounce word called chromothripsis.
'chromo-' = chromosome ; '-thripsis' = shattering into pieces
TL;DR at end (...)
nature.com/articles/s4158…
'chromo-' = chromosome ; '-thripsis' = shattering into pieces
TL;DR at end (...)
nature.com/articles/s4158…
The researchers' used short-read (~40X) whole-genome #sequencing on ~2,700 tumors across 38 cancer types (most tumors were advanced).
The goal was to study the frequency of a mutational phenomenon (chromothripsis), which previously was though to exist in only 2-3% of tumors.
The goal was to study the frequency of a mutational phenomenon (chromothripsis), which previously was though to exist in only 2-3% of tumors.
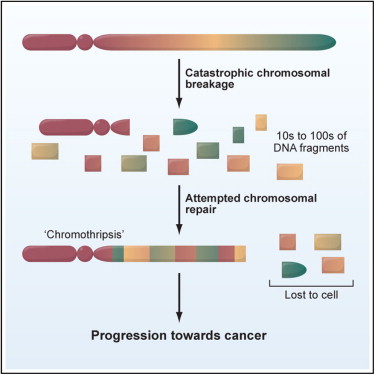
The prevailing view on tumor formation is that somatic mutations gradually accumulate over time, eventually overwhelming a cell's DNA repair machinery. Conversely, chromothripsis (above) is a single, catastrophic event defined by hundreds of structural rearrangements all at once.
Many previous studies of chromothripsis used arrays, which are not as sensitive to small copy-number variants (a defining feature of chromothripsis). My guess is that arrays systematically underrepresented the presence of chromothripsis in many tumors.
illumina.com/techniques/pop…
illumina.com/techniques/pop…
Using short-read #NGS, the researchers discovered a much greater prevalence of chromothripsis in a majority of human cancers (y-axis = % of tumors w/ chromothripsis). 

This may suggest that using long-read sequencing (which is even more sensitive to copy-number variation than short-read), will allow for even more precise detection of chromothripsis. Why is this potentially important in clinical #oncology?
ncbi.nlm.nih.gov/pmc/articles/P…
ncbi.nlm.nih.gov/pmc/articles/P…
Conceptually, chromothripsis falls under tumor mutational burden (TMB) - a well-studied biomarker used to indicate the use of targeted immunotherapies. If long-reads are even more sensitive to chromothripsis, we stand to boost the TMB detection and identify subgroups of patients-
-that may benefit from targeted therapies. It's likely that chromothripsis also may be used as a prognostic metric.
TL;DR:
Researchers sequenced tumors using sequencing, a more accurate technology, and discovered that a nasty mutational event (chromothripsis) was way more ...
TL;DR:
Researchers sequenced tumors using sequencing, a more accurate technology, and discovered that a nasty mutational event (chromothripsis) was way more ...
... common than previously thought. My guess is that using an even more accurate technology (long-reads) will find even more chromothripsis. This is important because chromothripsis is one reason doctors can prescribe immunotherapies.
• • •
Missing some Tweet in this thread? You can try to
force a refresh