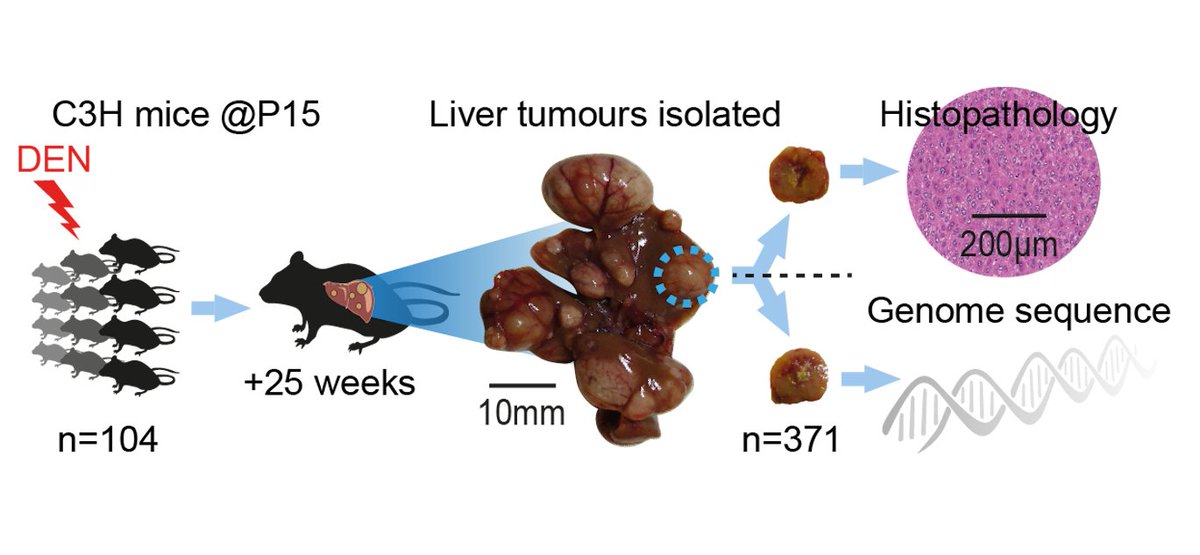
nature.com/articles/s4158…
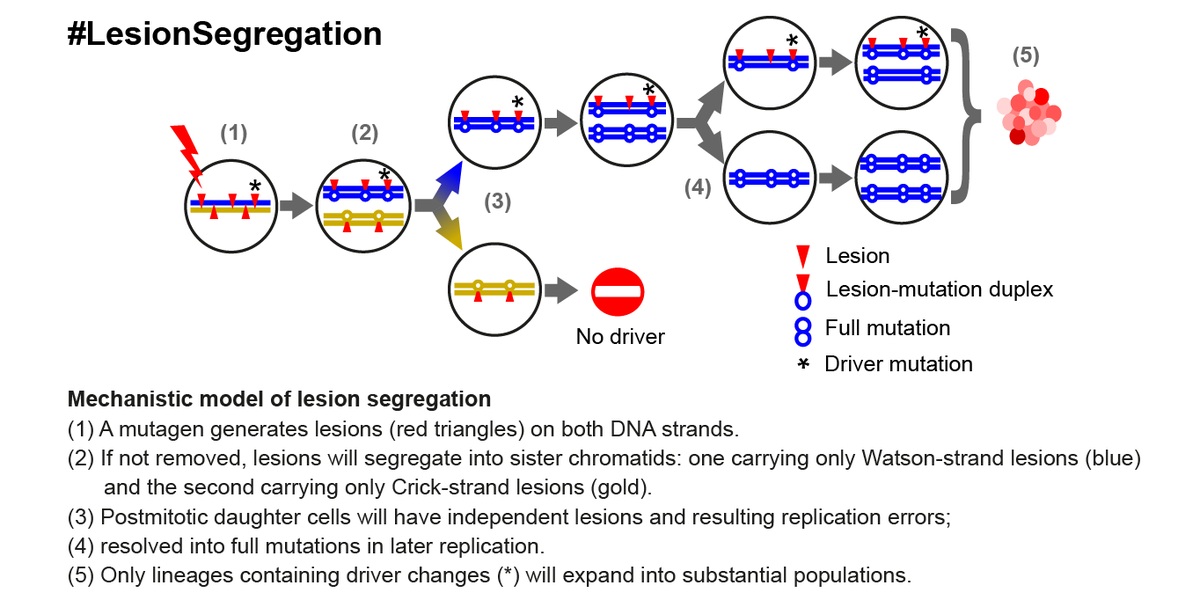
Here’s a #tweetorial to explain our #LesionSegregation findings...
[1/13]
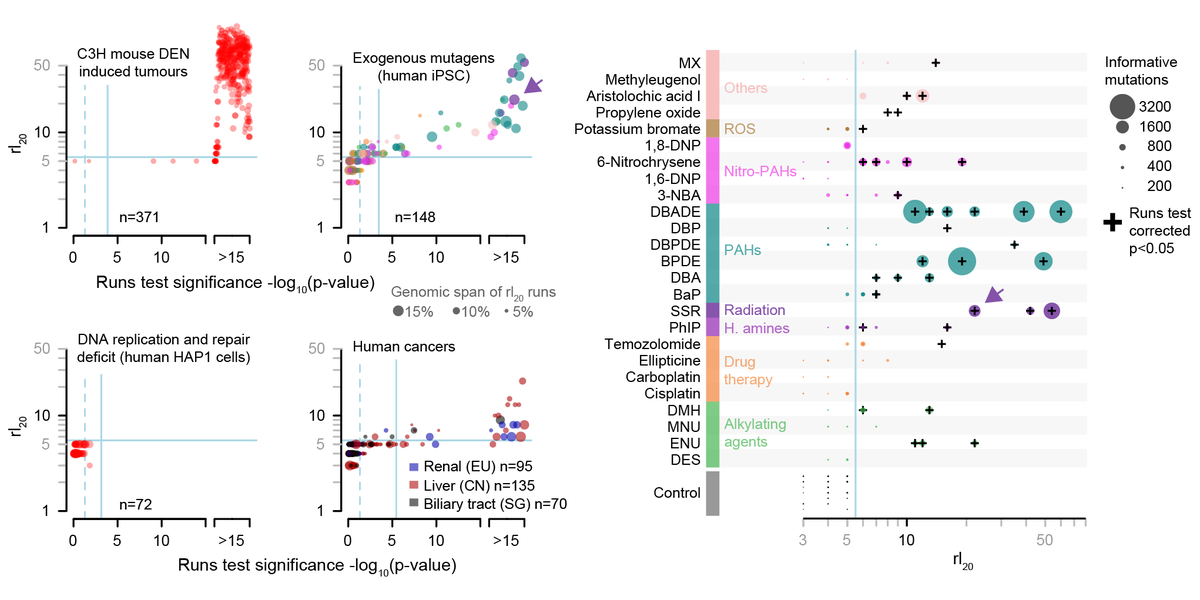
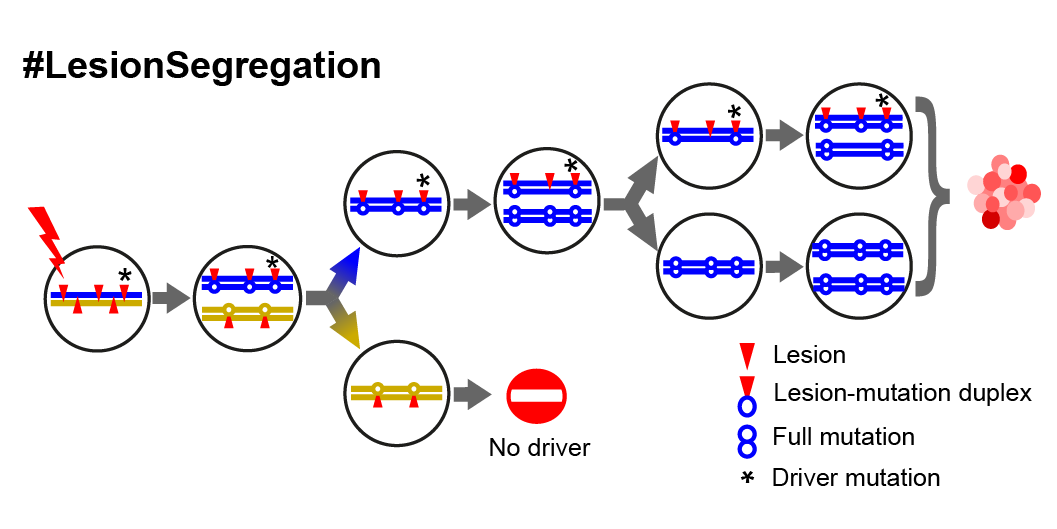
- Mutagens generate lesions on each DNA strand, and segregate during replication
- Daughter cells have non-overlapping lesions, which are then resolved into full mutations
- Lineages with drivers undergo clonal expansion
[5/13]
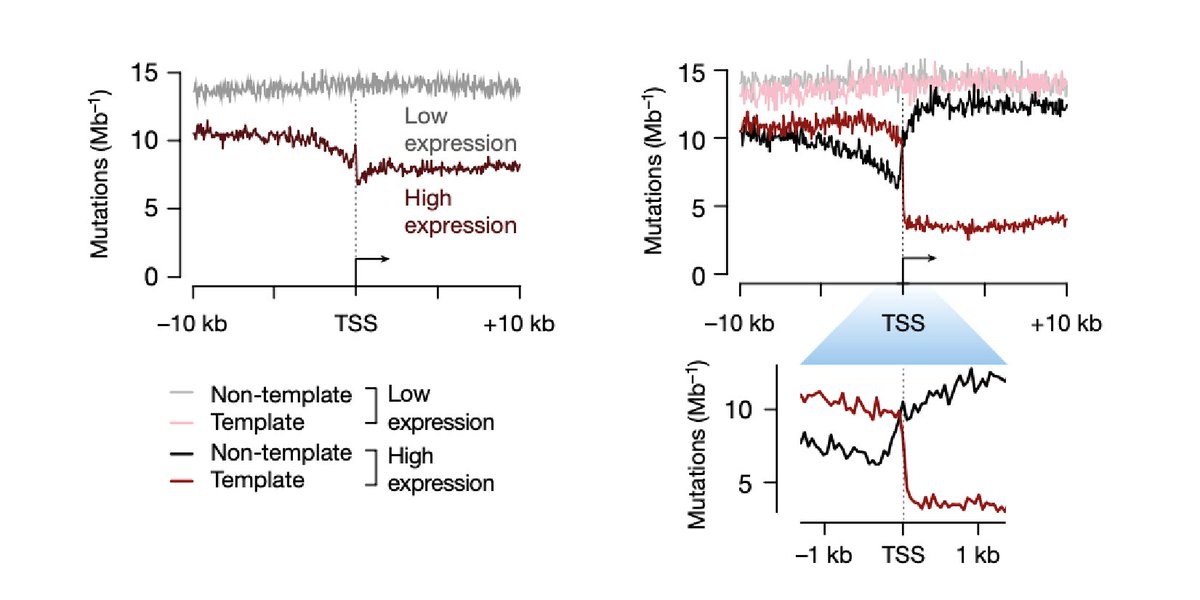
[6/13]
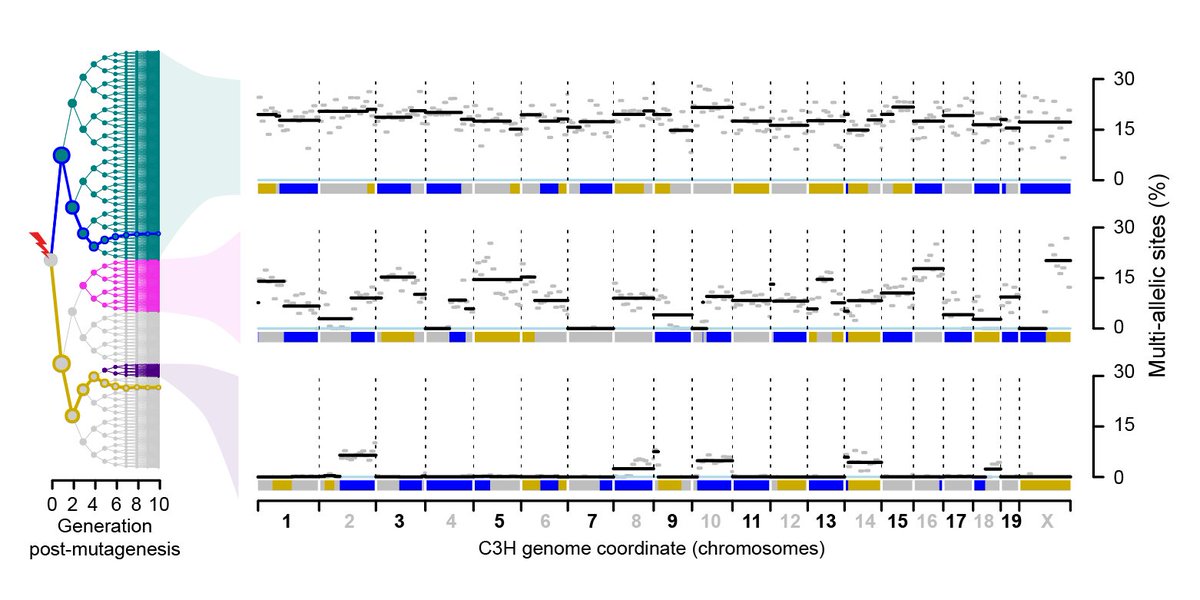
[7/13]
[8/13]
[9/13]
🇬🇧🇪🇸🇮🇳🇩🇪🇪🇪🇬🇷🇳🇱🇮🇪🇱🇻🇺🇸
[10/13]

[11/13]
And, of course, generous funding from @CR_UK, @ERC_Research, @wellcometrust, @EMBL, @The_MRC, @IRBBarcelona, @NIHRresearch.
[12/13]