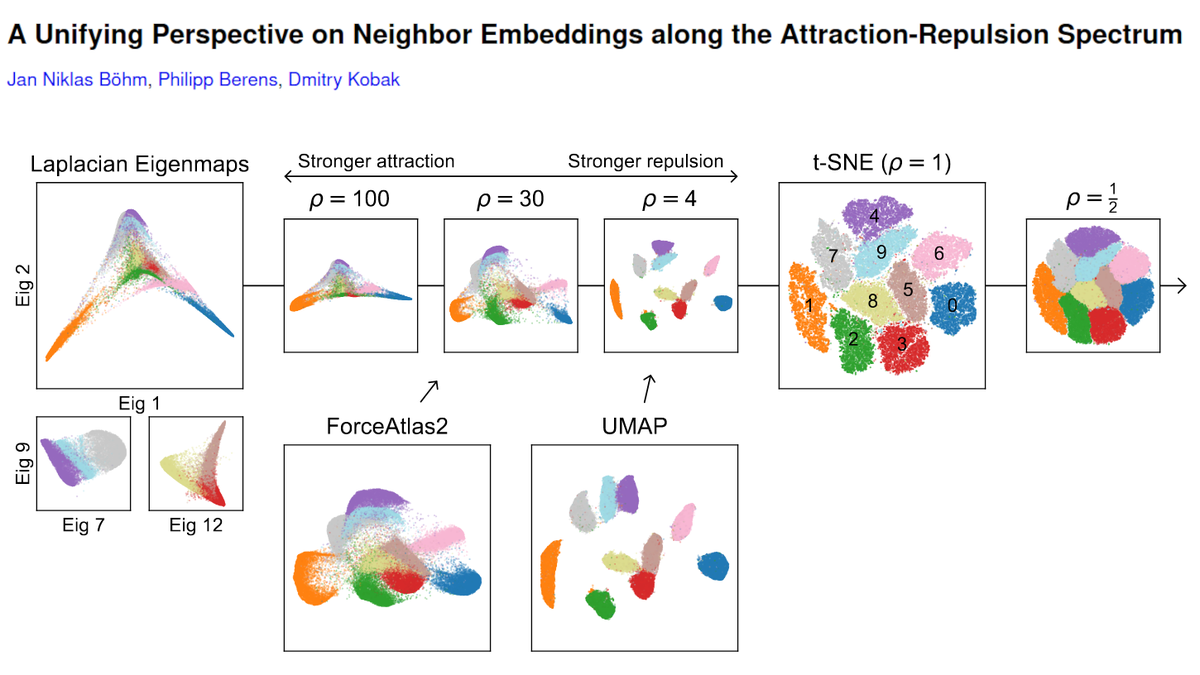
Remember the galaxy-like UMAP visualization of integers from 1 to 1,000,000 represented as prime factors, made by @jhnhw?
I did t-SNE of the same data, and figured out what the individual blobs are. Turns out, the swirly and spaghetti UMAP structures were artifacts :-(
[1/n]
I did t-SNE of the same data, and figured out what the individual blobs are. Turns out, the swirly and spaghetti UMAP structures were artifacts :-(
[1/n]

Here is the original tweet by @jhnhw. His write-up: johnhw.github.io/umap_primes/in…. UMAP preprint v2 by @leland_mcinnes et al. has a figure with 30,000,000 (!) integers.
But what are all the swirls and spaghetti?
Unexplained mystery since 2008. CC @ch402. [2/n]
But what are all the swirls and spaghetti?
Unexplained mystery since 2008. CC @ch402. [2/n]
https://twitter.com/jhnhw/status/1031829726757900288
The input here is a 1,000,000 x 78,628 matrix X with X_ij = 1 if integer i is divisible by the j'th prime number, and 0 otherwise. So columns correspond to 2, 3, 5, 7, 11, etc. The matrix is large but very sparse: only 0.0036% of entries are 1s. We'll use cosine similarity. [3/n] 

I use openTSNE by @pavlinpolicar. It uses Pynndescent by @leland_mcinnes to construct the kNN graph for sparse inputs. I'm using uniform affinities with k=15.
NB: t-SNE is faster than UMAP and needs less memory. I could run t-SNE *but not UMAP* on my 16Gb RAM laptop. [4/n]
NB: t-SNE is faster than UMAP and needs less memory. I could run t-SNE *but not UMAP* on my 16Gb RAM laptop. [4/n]

First of all -- the current version of UMAP does not produce any swirls/spaghetti 😧 I was getting them back in February (with UMAP 0.2 or 0.3), but with UMAP 0.4 I only get blobs and some doughnuts. This suggests that swirls/spaghetti were convergence artifacts. [5/n] 

t-SNE only shows blobs (and some stardust). Can we understand what they are?
Yes We Can!
The max number of prime factors (max row sum) in this dataset is 7. Coloring the embeddings by the number of prime factors shows that each blob has the same number of them. [6/n]
Yes We Can!
The max number of prime factors (max row sum) in this dataset is 7. Coloring the embeddings by the number of prime factors shows that each blob has the same number of them. [6/n]

Integers with two prime factors (orange) make up multiple blobs. Turns out, the largest blob consists of all such numbers that are divisible by 2. The next one --- of all such numbers divisible by 5. The next one -- by 7, etc. Here are labels up to 19. CC @wtgowers. [7/n] 

Similarly, numbers with three prime factors (green) are grouped into blobs by a combination of two shared prime factors. Etc.
This makes total sense if one thinks about how cosine similarity works. All numbers in one blob are equidistant from each other. [8/n]
This makes total sense if one thinks about how cosine similarity works. All numbers in one blob are equidistant from each other. [8/n]
So my guess is that the doughnuts in UMAP 0.4 are optimization artifacts of some kind: blobs do not really have any internal structure, as correctly shown by t-SNE. Why they appear in UMAP, I don't know.
For completeness, here are both UMAP versions labeled this way. [9/n]
For completeness, here are both UMAP versions labeled this way. [9/n]

This was a lot of fun to play around with. And led to several improvements in openTSNE along the way.
The main lesson, I guess, is that convergence artifacts can be very pretty ;-)
[10/10]
The main lesson, I guess, is that convergence artifacts can be very pretty ;-)
[10/10]
And now an animation! Gradually showing all integers from 1 to 1,000,000 in 50 steps of 20,000. [11/10]
• • •
Missing some Tweet in this thread? You can try to
force a refresh