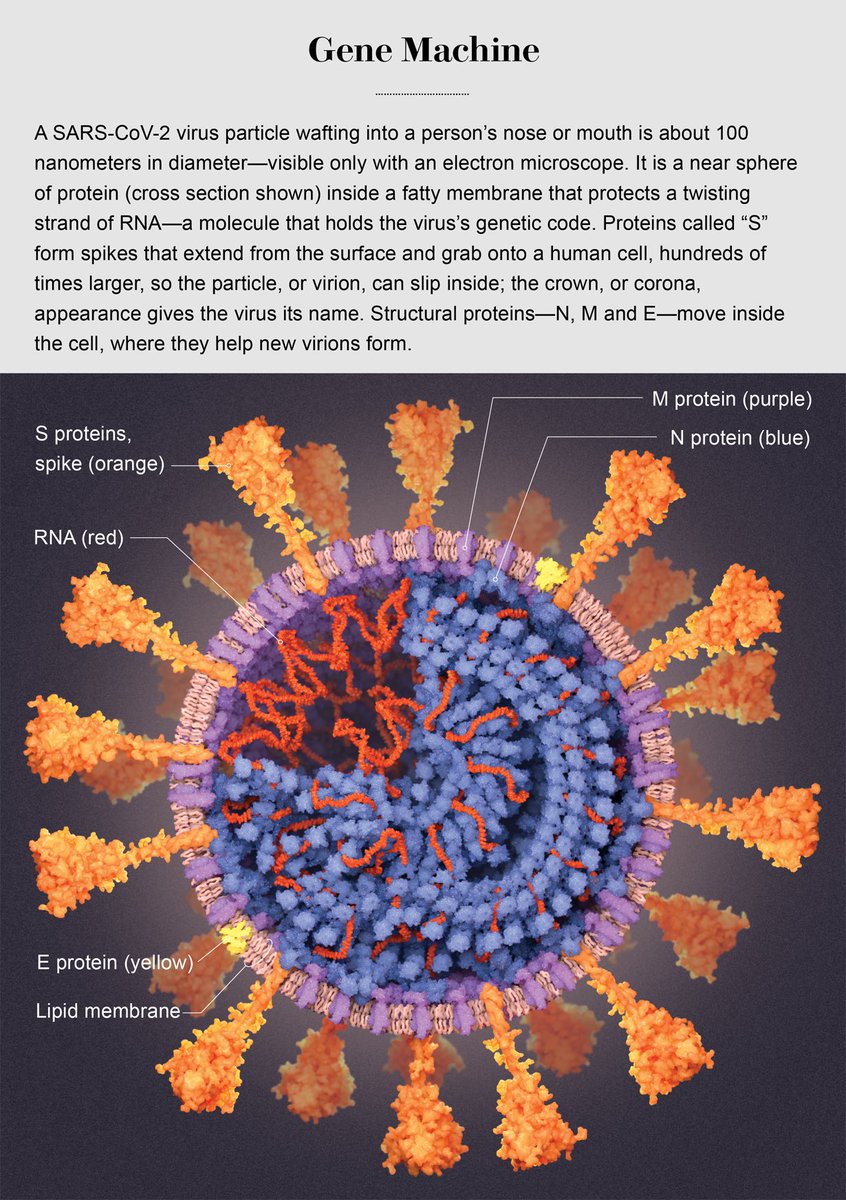
“if a similar phenomenon of host adaptation had occurred upon its jump into humans, those human-specific mutations would likely have reached fixation.. before the first SARS-CoV-2 genomes were generated.” 🙏🏻 @LucyvanDorp @BallouxFrancois biorxiv.org/content/10.110…
"The secondary host jump from humans into minks offers a glimpse into the window of early viral host adaptation of SARS-CoV-2 to a new host that has likely been missed at the start of the COVID-19 pandemic... and point to rapid adaptation of SARS-CoV-2 to a new host." 💯🔥
"pandemic is understood to have been caused by a unique host jump into humans from a single yet-undescribed zoonotic source in the latter half of 2019"
How does this fit with: "human-specific mutations would likely have reached fixation.. before the first SARS-CoV-2 genomes" 🤔
How does this fit with: "human-specific mutations would likely have reached fixation.. before the first SARS-CoV-2 genomes" 🤔
Is the hypothesis that a single natural spillover occurred, after which there was only 1 surviving lineage that led to COVID-19? All other adaptations to human died off or have not been detected? I guess China could've been that effective at wiping out all COVID in their country.
• • •
Missing some Tweet in this thread? You can try to
force a refresh