@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen What we know about the 7896 clade? Let's recap:
1/ They were published in June 2020 as part of a batch of 630 viruses of Latinne et. (2020). No details were given
ncbi.nlm.nih.gov/nuccore/?term=…
1/ They were published in June 2020 as part of a batch of 630 viruses of Latinne et. (2020). No details were given
ncbi.nlm.nih.gov/nuccore/?term=…
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen 2/ They are exactly 8 betas (the 8 remaining SARSr of the addendum?). Some are part of samples with not declared co-infections.
7896=7905=7907=7909=7921=7924, and 7931 with 2dS (at the end), and 7952 with those 2dS + 1 dS more
.
7896=7905=7907=7909=7921=7924, and 7931 with 2dS (at the end), and 7952 with those 2dS + 1 dS more
https://twitter.com/franciscodeasis/status/1308774597651767298
https://twitter.com/franciscodeasis/status/1309161872046907394
.
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen 3/ Surprisingly their names in Genbank do not include the "SARS-like" label
https://twitter.com/franciscodeasis/status/1309222654906335234
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen 4/ There is no clue in the paper or Genbank about where and when they were collected. But thanks to Bigd we know it was in Yunnan and in May-15 (the dates were tricked, btw)
.
https://twitter.com/franciscodeasis/status/1281272430045077505
https://twitter.com/franciscodeasis/status/1280812203453698048
.
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen 5/ Some amplicons of RaTG13 included a strange "7896" label in their filenames
https://twitter.com/franciscodeasis/status/1280518834965950472
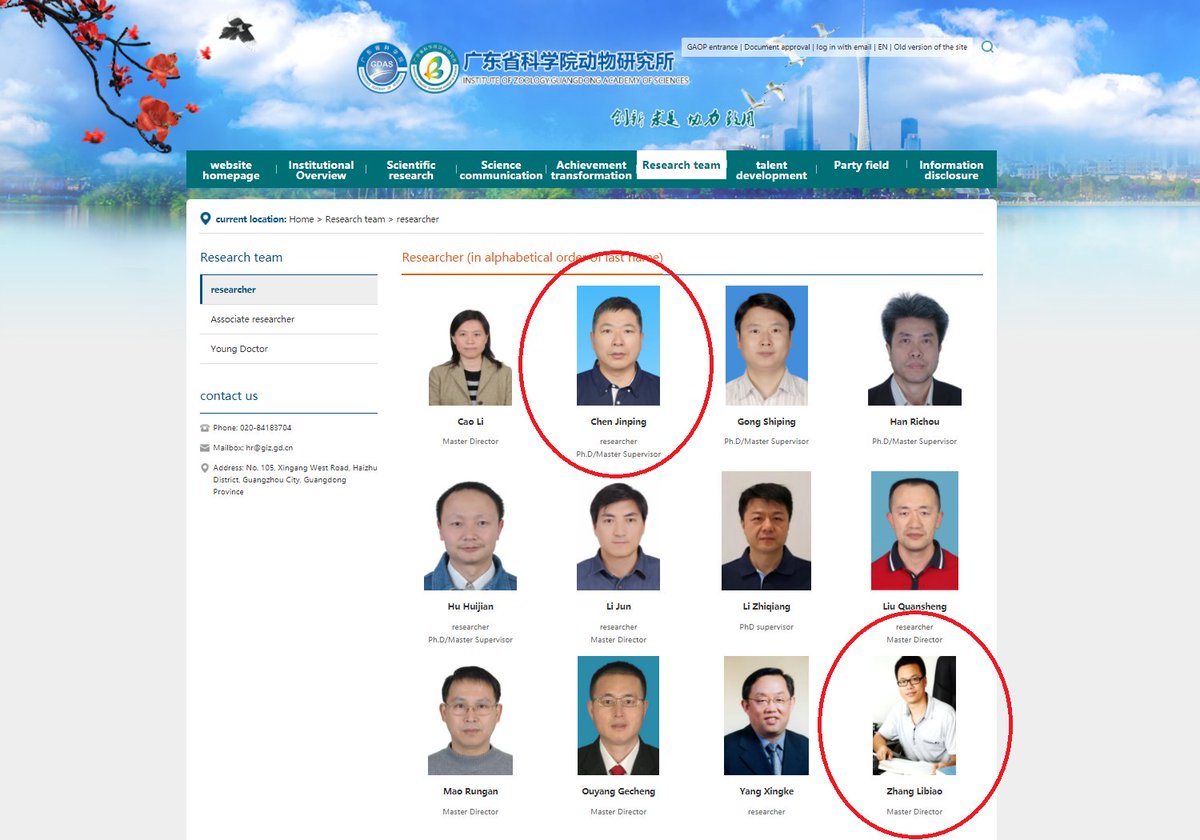
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen 6/ AFAIK, nobody has ever mentioned any of these viruses of the 7896 clade. Not even on a phylogenetic tree. Just only @babarlelephant
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant 7/ Could it be that those 8 SARSr viruses were collected in TG mineshaft? Yes, there is some evidence pointing to that:
- Types of co-infections:
- Timing: 8130 is probably from Jinning
- Bat species:
Also:
- Types of co-infections:
https://twitter.com/franciscodeasis/status/1309151461939589120
- Timing: 8130 is probably from Jinning
- Bat species:
https://twitter.com/franciscodeasis/status/1285983116356653061
Also:
https://twitter.com/franciscodeasis/status/1289713229510463489
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant 8/ Some possible derived hypothesis:
- SL3 group:
- WIV15:
- PREDICT involvement:
.
- SL3 group:
https://twitter.com/franciscodeasis/status/1288513072781504513
- WIV15:
https://twitter.com/franciscodeasis/status/1280826716659859456
- PREDICT involvement:
https://twitter.com/franciscodeasis/status/1280647559917297664
.
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant End/ This 7896 virus looks important because it was the cause of being blocked by @PeterDaszak
https://twitter.com/franciscodeasis/status/1280980860850909189
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak Addendum:
A1/ The clade 7896 is totally irrelevant in Latinne et al. (2020) despite being the 2nd in the line of succession of SARS-CoV-2 just after 4991 (or the 3rd just after RmYN02 if we take it as valid). There is only the accession number in a big table in Supplementary data
A1/ The clade 7896 is totally irrelevant in Latinne et al. (2020) despite being the 2nd in the line of succession of SARS-CoV-2 just after 4991 (or the 3rd just after RmYN02 if we take it as valid). There is only the accession number in a big table in Supplementary data
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A2/ Latinne et al. (2020) paper shows a bad inventory management of samples in WIV. Among the samples of the paper there are several duplicates, conflicts of IDs, inconsistencies and reposts
https://twitter.com/franciscodeasis/status/1272638480401731586
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A2 / Some other examples:
-
-
-
And in Bigd the samples of Laos are not included:
-
https://twitter.com/franciscodeasis/status/1272182561645699072
-
https://twitter.com/franciscodeasis/status/1283438622281216011
-
https://twitter.com/franciscodeasis/status/1293697477170929664
And in Bigd the samples of Laos are not included:
https://twitter.com/franciscodeasis/status/1281267507479920645
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A3/ Genbank "submitted date" for 7896 was 13-AUG-2019
ncbi.nlm.nih.gov/nuccore/MN3126…
ncbi.nlm.nih.gov/nuccore/MN3126…
https://twitter.com/franciscodeasis/status/1283536162171293696
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A4/ RaTG13 amplicons dates included some of 2017. And all those amplicons have also the "7896" label. In addition, the WIV has now recognized earlier sequencing of RaTG13, but in 2018, not in 2017. So, what is the meaning of that 2017?
https://twitter.com/franciscodeasis/status/1281364285399224320
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A5/ The 6 betas of the 7896 clade that are identical had their RdRp sequenced longer than usual. The other 2 betas, with some mutations, were normally sequenced.
Note: I used this query, downloaded, and created a pivot table
ncbi.nlm.nih.gov/nuccore/?term=…
Note: I used this query, downloaded, and created a pivot table
ncbi.nlm.nih.gov/nuccore/?term=…

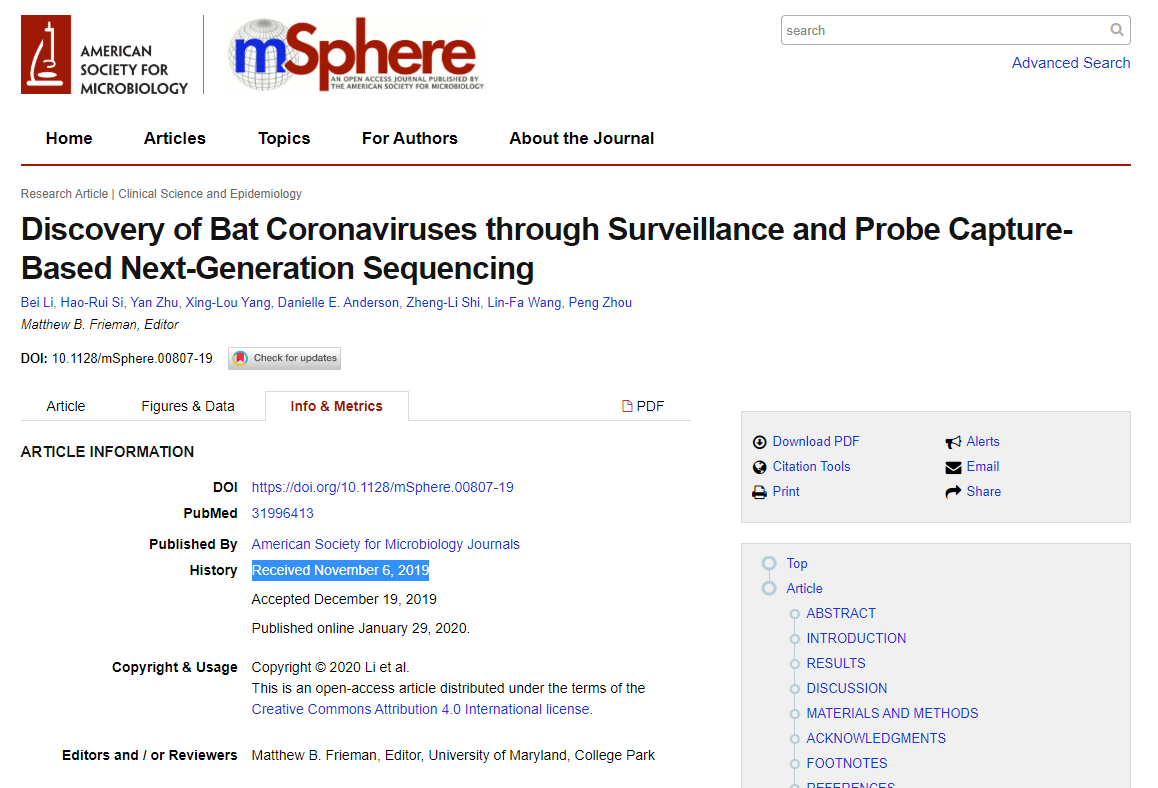
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A6/ Latinne et al. (2020) was submitted to @nature for review on October 6th, 2019.
There is a pre-print on May 31st, 2020, which is essentially the same as the final paper, but there must be a previous version for obvious reasons
nature.com/articles/s4146…
biorxiv.org/content/10.110…

There is a pre-print on May 31st, 2020, which is essentially the same as the final paper, but there must be a previous version for obvious reasons
nature.com/articles/s4146…
biorxiv.org/content/10.110…


@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A6/ No need to explain that this first draft of Latinne et al. (2020) must be disclosed and opened for public scrutiny
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A7/ This tweet, with a phylogenetic tree shown by ZLS after the publication of this thread, cannot be missing in this thread
https://twitter.com/emmecola/status/1334505779718729729
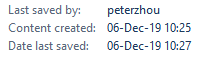
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A8/ And another thing that Nature must clarify: Why accessions were created one month later than the paper was sent for peer review (06-Oct-19)?
Note: Genbank IDs are created [supposedly] within 2 days from submission to NCBI
Note: Genbank IDs are created [supposedly] within 2 days from submission to NCBI
https://twitter.com/franciscodeasis/status/1357110425108828161
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A9/ Something we cannot forget when talking about the 7896 clade, because it is certainly incredible
https://twitter.com/franciscodeasis/status/1357111003041923072
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A10/ Why the "submitted" date of these 7896 clade (and the others samples of Latinne et al. (2020) is 13-AUG-19?
@nature must know what happened and must clarify too!
@nature must know what happened and must clarify too!
https://twitter.com/franciscodeasis/status/1359157337630646277
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A11/ BIGD corrected the dates in the exact way we said some months ago. They probably obfuscated the dates not only for hiding sampling many days in a row, but also to make it more difficult to cluster the samples and figure out the number of locations
https://twitter.com/franciscodeasis/status/1356980344445100033
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A11/ Dates in BIGD are very strange. "Release" is supposed to happen after "submission".
BioProject:
- Release date: 2020-04-11
- Submission date: 2020-04-09
Beta 7896:
- Release date: 2020-04-11
- Submission date: 2020-06-03
(Note: in Genbank 01-Jun-20)
bigd.big.ac.cn/biosample/brow…
BioProject:
- Release date: 2020-04-11
- Submission date: 2020-04-09
Beta 7896:
- Release date: 2020-04-11
- Submission date: 2020-06-03
(Note: in Genbank 01-Jun-20)
bigd.big.ac.cn/biosample/brow…
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A12/ Did they continue going to the mine after this last visit in which they collected this clade?
As we saw, they seems to be commited by the article to publish 7896 and concede visits until 2015. But in 2018 there was walking activity in the mine
As we saw, they seems to be commited by the article to publish 7896 and concede visits until 2015. But in 2018 there was walking activity in the mine
https://twitter.com/franciscodeasis/status/1359098532561625089
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A13/ BOOM:
Accession MN312671 (beta 7896) was created on November 7th, 2019 (as I predicted using Genbank ID method).
More interesting: two updates during the embargo and after the outbreak, with no traces in the [public] revision history
ncbi.nlm.nih.gov/nuccore/MN3126…
Accession MN312671 (beta 7896) was created on November 7th, 2019 (as I predicted using Genbank ID method).
More interesting: two updates during the embargo and after the outbreak, with no traces in the [public] revision history
ncbi.nlm.nih.gov/nuccore/MN3126…

@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak Here NCBI answer. Wish I knew the meaning of "insignificant" here 

@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak A14/ Another BOOM
As we already deduced in November, this slide from Zhengli Shi showed in December proves that the 7896-clade was collected in the mine!
Thanks @Rossana38510044 for finding it!
And another scandal: they have the spikes unpublished!
vimeo.com/490671967 (14:41)
As we already deduced in November, this slide from Zhengli Shi showed in December proves that the 7896-clade was collected in the mine!
Thanks @Rossana38510044 for finding it!
And another scandal: they have the spikes unpublished!
vimeo.com/490671967 (14:41)

@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak And this is very suspicious: why they "adapted" that slide from Hu et al. (2017) and not from Cui et al. (2019)? Want to redefine clade 2 to hide that "SL3" (clade 3) was sequenced in 2017?
Btw, note that 7896-labelled amplicons were dated as 2017!
Btw, note that 7896-labelled amplicons were dated as 2017!
https://twitter.com/franciscodeasis/status/1288513072781504513
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak Also thanks for @TheSeeker for finding the reference of Ra7909 and noting the fact of being sequenced more than just its RdRp (all that has been published until now)
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak @TheSeeker And @DecrolyE for posting the webinar
https://twitter.com/DecrolyE/status/1365724145741402113
@AntGDuarte @Rossana38510044 @jhouse678 @nature @edwardcholmes @arambaut @angie_rasmussen @K_G_Andersen @babarlelephant @PeterDaszak Why Ra7909 and Ra7924 in clade 1 if they have deletions in the RBD?
The criteria for classification in clades was the size vs SARS1:
nature.com/articles/s4157…
They probably created a 3rd clade for them. "SL3", as the filename of the metagenome of RaTG13

The criteria for classification in clades was the size vs SARS1:
nature.com/articles/s4157…
They probably created a 3rd clade for them. "SL3", as the filename of the metagenome of RaTG13
https://twitter.com/franciscodeasis/status/1285386277039288320


• • •
Missing some Tweet in this thread? You can try to
force a refresh