
Opening the tech update at the Nanopore Community Meeting, from Oxford @Nanopore, James Clark, VP platform technology #nanoporeconf
JC: a reminder that our goal is to enable the analysis of anything, by anyone, anywhere #nanoporeconf
JC: Looking at about the key qualities of nanopore sequencing: high accuracy data, with high yields, information-rich data, a versatile platform that scales from portable to ultra-high throughput #nanoporeconf
JC: a reminder: this is how nanopore sequencing works. The current trace, called a squiggle, is decoded by basecalling into a sequence vimeo.com/337258910 #nanoporeconf
JC: you can see previous videos, from Oxford Nanopore or the community, at our resource centre nanoporetech.com/resource-centre
JC: this is the range of @nanopore devices – all sizes mean that anyone can access the technology however big the project. PromethION, GridION and MinION Mk1C include embedded compute #nanoporeconf. More here nanoporetech.com/products 

JC: PromethION is our largest device, designed for larger samples and larger projects #nanoporeconf
JC: at London Calling we reviewed some PromethION flow cell improvements, in connectivity to the ASIC and microwell structure. More recently we’ve been shipping FCs that include these improvements & are seeing rising data yields from Prom flow cells in the field #nanoporeconf
JC: we perform PromethION benchmarking tests regularly, and in June 2020 we achieved 8Tb in a single run using PCRd DNA, on all 48 flow cells (~170Gb avg per FC). #nanoporeconf
JC: one of the elements of the technology is the membrane that the nanopores are embedded in. We have redesigned a membrane that enables a higher number of pores in channel, which translates to higher throughput. This is now rolled out #nanoporeconf
JC: As a result of these improvements we have a new internal record of *10 Terabases* of data from one run of all 48 PromethION flow cells, averaging 208 Gb per flow cell – with excellent consistency between flow cells #nanoporeconf
JC: the 10Tb record utilises the latest MinKNOW, membrane, and used PCRd human DNA prepped on Hamilton #nanoporeconf
JC: A new ultra-long kit which has been optimised for people interested in those valuable >100kb fragments. Internally we can produce 90-100Gb (read N50>100kb) data from a PromethION FC, with a new record of 4.15Mb single read #nanoporeconf 

JC: We are now achieving internally 220Gb per PromethION FC with 10-30kb N50, or 100Gb per FC at 100kb – this requires some interaction during a run. When barcoded, => 2 human genomes at 30X or trio at 20X #nanoporeconf 

JC: we're delighted to see users' yields climbing and we look forward to supporting you, building more on this success
https://twitter.com/AW_NGS/status/1331243073553174528
JC: looking at addressing limitations for future yield improvements, we are working on increasing the concentration of mediator in the well and how we can increase channel numbers #nanoporeconf
JC: Moving onto the smallest nanopore flow cell: Flongle. This is designed to enable single, rapid, on-demand analyses at low cost. #nanoporeconf
JC: for Flongle we think about how to drive flow cell costs even lower, to make it possible for more people to use it to run more analyses. This is all about manufacturing innovation: not just flow cell design but better processes for making them at scale #nanoporeconf
JC: Whats the pathway to Flongle being very low cost? Nothing in the flow cell needs to be expensive – here is a prototype single cell channel from back in 2006 in which we successfully ran single pores. #nanoporeconf 

Talking of manufacturing innovation - here is the high tech factory we opened in 2019
JC: For low cost Flongle flow cells, we can replace a metal base with a plastic sheet. We can go from lithographic processes to cheaper mouldings. We want to move towards flow cells for just a few dollars #nanoporeconf 

JC: We continue to work on voltage sensing; a long running project that is high risk and very high reward – to dramatically increase density of nanopore sensing. Lots of progress and the project is starting to look less risky #nanoporeconf
JC: on voltage sensing chip - if these channels are engineered at a 40um pitch, this has the potential to deliver ~100,000 nanopore channels on a piece of silicon 16x30mm => 3.9Tb / day #nanoporeconf 
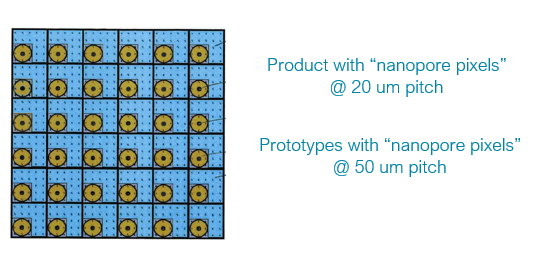
Thanks to James Clarke for part 1 of today's update - up next, Stuart Reid, VP Development #nanoporeconf
SR: where does the high accuracy of nanopore sequencing data come from? There is a rich signal - patterns to be learned as the DNA travels through the nanopore. A pretty good application of machine learning #nanoporeconf
SR: Different generations of algorithms, on the same chemistry, have moved raw read accuracy of *each single, native molecule* from ~90% to >98% in around 2 years. #nanoporeconf
SR: Research version of bonito, delivering modal single-read accuracy of ~98.3% is available now: github.com/nanoporetech/b… – but it’s ready for prime time – expect to see in Guppy and MinKNOW in coming months #nanoporeconf
SR: high-quality consensus accuracies now achievable with bonito using only half the read depth – R9.4.1 hits Q45 at 30-60X, and R10.3 gets to Q50 at less than 100X #nanoporeconf
SR: combination of Medaka with DeepVariant - similar to community methods - have boosted SNP calling to ~99.92%, expect further improvement with new basecallers #nanoporeconf
SR: new SV calling pipeline makes uses of excellent Community tools LRA @mjpchaisson and cuteSV to produce high-quality calls twice as fast and with half as much data #nanoporeconf
SR: new ultra-long kit achieves 90-100 Gb of data per PromethION, with hundreds of thousands of reads >100 kb, and hundreds of reads >1 Mb; will be released shortly. Ideal for spanning complex SVs, spanning repeats, resolving regions challenging to assemble @circulomics
SR: re-basecalling T2T consortium CHM13 data with bonito gives 80 Mb N50, Q46, BUSCO 96%. Pore-C can improve further with long-range contact information – see Poster section for more #nanoporeconf
SR: new PCR cDNA kit in early access now; improvements give ~100 million reads per PromethION flow cell with >85% full-length & mapped, to deliver unprecedented ability to quantify isoform expression levels #nanoporeconf
SR: reminder – only @nanopore can directly examine modified bases, with no GC bias, long read lengths, phasing etc – it’s all in the data. R10.3 shows high sensitivity to modifications, delivering even more accurate per-read methylation calling #nanoporeconf
SR: R9.4.1 is good at methylation calling - an F1 score of 91.5% per read - but R10.3 is an improvement at 95% #nanoporeconf
SR reviews current levels of different types of accuracy - more information can be found here nanoporetech.com/accuracy #nanoporeconf
SR: you don't necessarily need to basecall to do Adaptive Sampling. A user can programme their system to accept or reject strands based on a configuration specified in software, enabling rapid sequencing of regions of interest, no sample prep
SR: a grand challenge @nanopore: make bioinformatics easy. EPI2ME Labs is the simplest way to explore bioinformatics with nanopore #nanoporeconf 

SR: The MinKNOW App is now available for download on iOS and Android devices, more information and download links here: nanoporetech.com/about-us/news/… #nanoporeconf 

SR: This week Oxford Nanopore has generated modal *raw-read accuracy* of 99.1% (99%=Q20) using a new chemistry with Bonito, delivered on internal validation sets with a substantial fraction of these raw reads above Q20 #nanoporeconf 
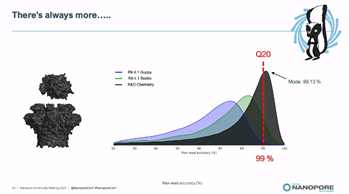
Next up @RosemaryDokos who is going to update us on the usability and accessibility of our technology; critical to support the goal of enabling the analysis of anything, by anyone, anywhere #nanoporeconf
RD: We focus on all parts of the workflow – versatile and easy sample prep, sequencing and analysis, including open communities #nanoporeconf 

RD: on preparation, its important to have the right tools. We have native DNA preparation methods and will be adding the ultra-long kit to this. And there are extensive options on the amplified side, & direct RNA – as well as ways of targeting regions of interest #nanoporeconf 

RD: we wrap the preparation methods with protocols, and have just updated the knowledge section of the nanopore community. Please give us feedback on the new protocol layouts #nanoporeconf
community.nanoporetech.com/docs/
community.nanoporetech.com/docs/
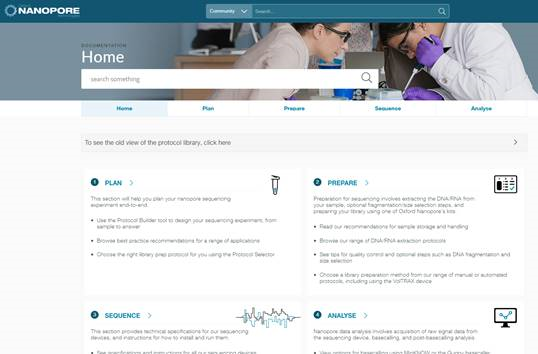
RD: EPI2ME labs has notebooks explaining how to process and manipulate nanopore data. GitHub has all our pipelines in open source. EPI2ME labs has now been available for six months #nanoporeconf
RD: over the coming months we’ll be working to take the workflows from EPI2ME labs into EPI2ME, so we can work towards a single click bioinformatics solution #nanoporeconf
RD celebrates the new working environments for biologists #nanoporeconf 

RD: Oxford Nanopore is collaborating with Hamilton to automate high throughput library preparation workflows on Hamilton’s NGS STAR 96. #nanoporeconf
RD: on Hamilton prep, 1st protocol enables preparation of 96 samples in under 5 hours, with only one hour of hands-on time. One run fills two PromethION 48s, enabling high yield, highly validated sample prep and sequencing output. #nanoporeconf
RD: we’re also working with Opentrons to deliver mid-range lab automation suitable for MinION/GridION. The robots are accessible, compact, & can support 2,500 LamPORE tests/day or automation of the Ligation kit, ARTIC native barcoding, & suitable for future PCR based applications
RD: we can’t talk about automation without mentioning VolTRAX – key to accessibility
RD: we’ve been completing all the validations for the PCR ready cartridge, including enabling RT-PCR, and are deploying it for ARTIC whole genome sequencing of #SARSCoV2; prepare on VolTRAX, sequencing on Flongle for $75 #nanoporeconf
RD: whats available in store for VolTRAX #nanoporeconf 

RD: Three configurations of GridION are now available, for research, Q-line for applied markets, and CE-IVD for diagnostic use #nanoporeconf 

RD: We make devices available with no capital requirements. As soon as you invest in us, we invest back, with a seamless upgrade pathway and no hefty renewal fees. The same users who were generating 1Gb of data on MinION a few years ago are now generating tens of Gb #nanoporeconf
RD: the full range of specs of nanopore products nanoporetech.com/products/compa…
RD: with high throughput projects on PromethION, there are choices of workflows, all delivering cost competitive, high value genomes #nanoporeconf 

RD: PromethION’s capital-free model delivers not only value, but huge capacity to deliver whatever volumes of data are required #nanoporeconf 
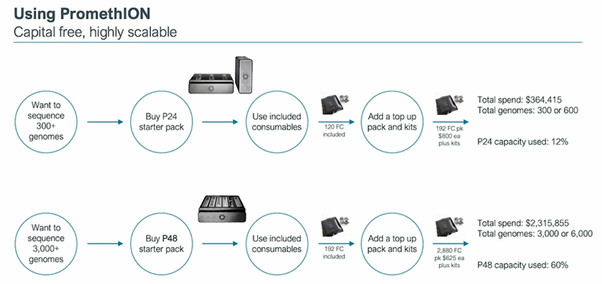
RD: Nanopore data is information-rich, so the data that you generate has additional value #nanoporeconf 

To see the whole talk from @RosemaryDokos, James Clarke and Stu Reid’s, visit #nanoporeconf (register nanoporetech.com/ncm20 if you aren't in the conference platform already) to watch the talk on demand and we'll make it available at our website soon #nanoporeconf
@threadreaderapp unroll!
• • •
Missing some Tweet in this thread? You can try to
force a refresh


