Congrats Nick github.com/Nick-Eagles for your @biorxivpreprint first pre-print! 🙌🏽
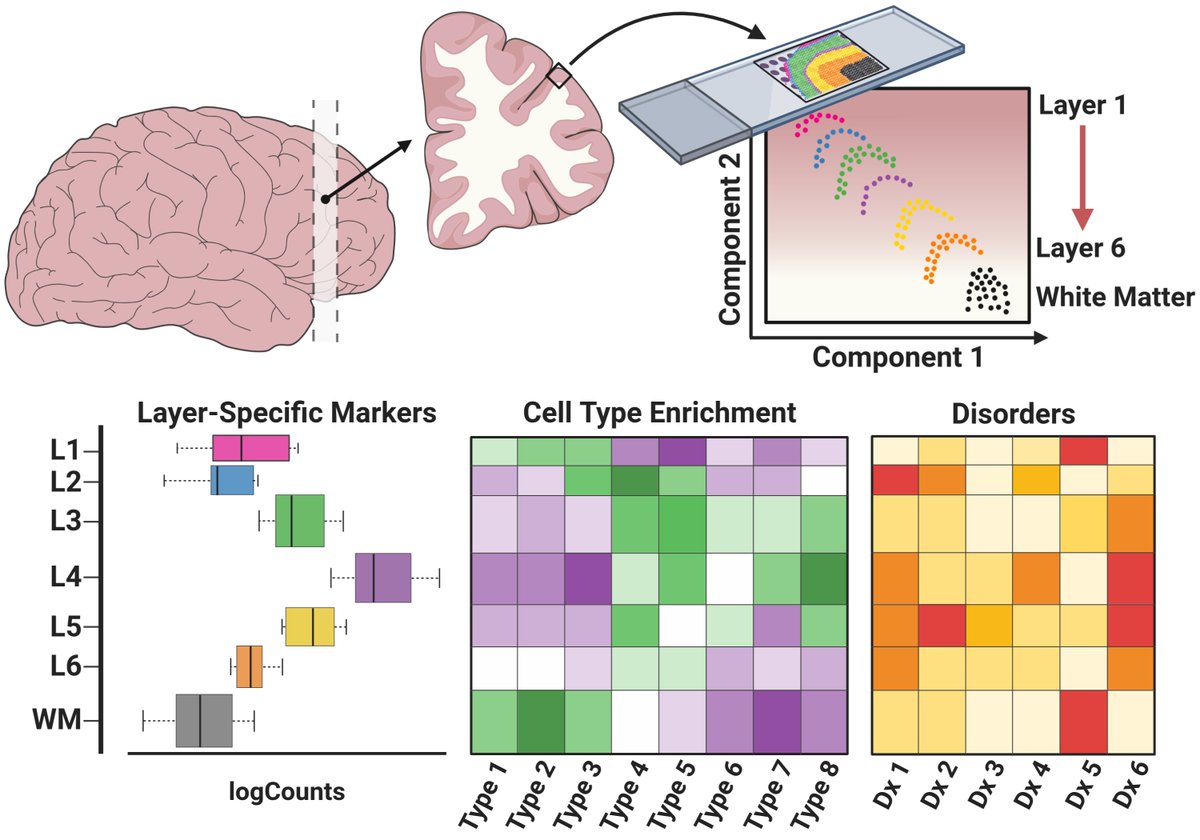
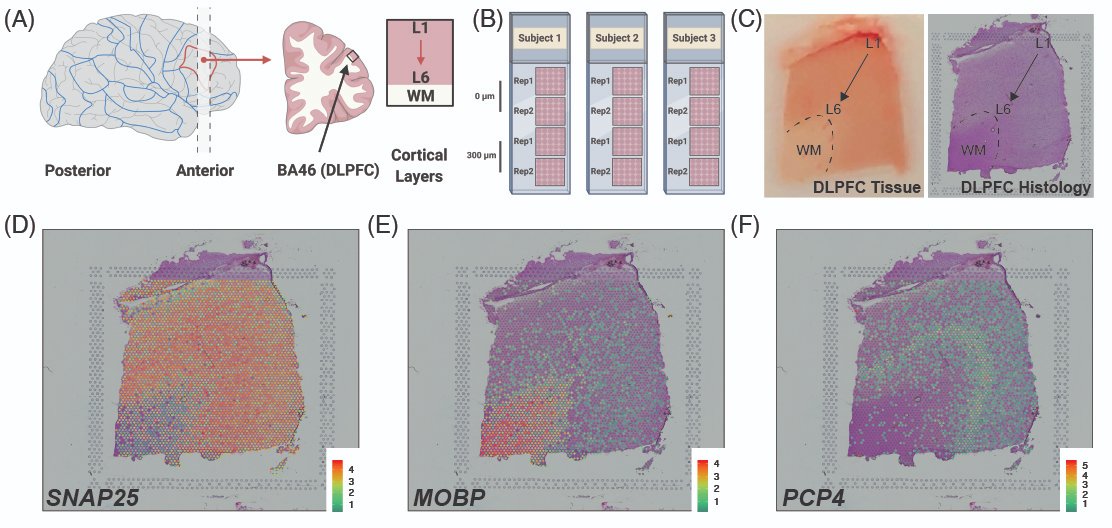
SPEAQeasy is our @nextflowio implementation of the #RNAseq processing pipeline that produces @Bioconductor-friendly #rstats objects that we use at @LieberInstitute
📜 biorxiv.org/content/10.110…
SPEAQeasy is our @nextflowio implementation of the #RNAseq processing pipeline that produces @Bioconductor-friendly #rstats objects that we use at @LieberInstitute
📜 biorxiv.org/content/10.110…

This project started a few years ago with Emily Burke linkedin.com/in/emilyeburke/ @BadoiPhan @andrewejaffe with whom we wrote a version specific to our HPC 🖥️ (JHPCE)
github.com/LieberInstitut…
Then @Jake_Aaron96 Israel Aguilar et al from @wintergenomics rewrote using it @nextflowio
github.com/LieberInstitut…
Then @Jake_Aaron96 Israel Aguilar et al from @wintergenomics rewrote using it @nextflowio

Nick continued developing it with input from @sudo_BreeB and tests from colleagues
Nick also wrote the documentation website research.libd.org/SPEAQeasy/
Then @JoshStolz2 @lahuuki helped show the outputs can be analyzed with @Bioconductor
research.libd.org/SPEAQeasy-exam…
Nick also wrote the documentation website research.libd.org/SPEAQeasy/
Then @JoshStolz2 @lahuuki helped show the outputs can be analyzed with @Bioconductor
research.libd.org/SPEAQeasy-exam…

We've used these #RNAseq outputs extensively in previous projects with @andrewejaffe et al
Some examples:
doi.org/10.1016/j.neur…
doi.org/10.1038/s41467…
Now our colleagues can use it too without having to tweak all the processing steps ^^
Some examples:
doi.org/10.1016/j.neur…
doi.org/10.1038/s41467…
Now our colleagues can use it too without having to tweak all the processing steps ^^
Next up, Nick & @JoshStolz2 will teach a workshop on how to use it at #EuroBioc2020 next week
eurobioc2020.bioconductor.org/conference_sch…
research.libd.org/SPEAQeasyWorks…
I also bet that you'll see Nick's name around more frequently. He's just getting started ;)
The first paper is always the hardest 😤
eurobioc2020.bioconductor.org/conference_sch…
research.libd.org/SPEAQeasyWorks…
I also bet that you'll see Nick's name around more frequently. He's just getting started ;)
The first paper is always the hardest 😤
• • •
Missing some Tweet in this thread? You can try to
force a refresh