As is becoming increasingly common these days, the pace of reality has well outstripped that of the scientific publication cycle. Still, I'd like to share this with you: sciencedirect.com/science/articl… (1/14)
Unless you've been entirely disconnected for the last month or three, the core message (that immunity-escaping variants of SARS-CoV-2 are developing and need to be watched) should come as no surprise. This work focuses on one of the first such mutants identified, N439K. (2/14)
At the time of submission, the N439K variant had been spotted in 34 different countries and had arisen independently multiple times, suggesting at least some gain of fitness over the wild-type virus. (3/14)
So, what does this "N439K" mean? It's a so-called "point" mutant - a change in the identity of a single amino acid - at residue 439 of the spike protein, from asparagine (a short, neutral-but-polar residue) to lysine (longer and positively charged). (4/14)
This site is important for at least two reasons:
(1) It's in the part of the spike protein receptor binding domain (RBD) that binds to ACE2, and is the target for so-called "neutralising" antibodies - the best kind, those that block the virus from actually infecting cells (5/14)
(1) It's in the part of the spike protein receptor binding domain (RBD) that binds to ACE2, and is the target for so-called "neutralising" antibodies - the best kind, those that block the virus from actually infecting cells (5/14)
... rather than "just" alerting the immune system to destroy the viral particle; and
(2) the equivalent site in the original SARS spike protein is an arginine (another positively-charged residue) which forms a stabilising salt bridge to glutamic acid 329 on ACE2.
(6/14)
(2) the equivalent site in the original SARS spike protein is an arginine (another positively-charged residue) which forms a stabilising salt bridge to glutamic acid 329 on ACE2.
(6/14)
So the questions were: (a) does N439K form a similar stabilising salt bridge, increasing affinity to ACE2; and (b) does this mutant escape recognition by existing antibodies?
The answers were (a) sorta, and (b) sometimes.
(7/14)
The answers were (a) sorta, and (b) sometimes.
(7/14)
To go into more detail: while the lysine doesn't appear to form a *direct* salt-bridge (where its amine binds directly to the acid), there is salt-bridging behaviour leading to a tighter RBD-ACE2 interface. Wild-type (6m0j) on left, N439K on right. (8/14) 



Experiments showed that this mutant has about 2-fold higher affinity for ACE2 compared to wild-type, and comparable overall infection fitness.
Now, immune escape: of a panel of 140 monoclonal anti-RBD antibodies, 16.7% had at least a 2-fold reduction in affinity. (9/14)
Now, immune escape: of a panel of 140 monoclonal anti-RBD antibodies, 16.7% had at least a 2-fold reduction in affinity. (9/14)
Not terrible, you might think, but the affected antibodies include some that are currently in clinical use for treatment of severe COVID-19 cases.
(10/14)
(10/14)
Overall, this mutant served as one of the early warning signs that the fight against SARS-CoV-2 could become even more protracted than it already has been, and that continual surveillance and adaptation will be needed. (11/14)
This has of course been borne out by the various other "problem" strains that have arisen since. Seems there's still a lot of work to do.
This paper was the work of a great many highly talented people, and I am grateful to have been given the chance to contribute. (12/14)
This paper was the work of a great many highly talented people, and I am grateful to have been given the chance to contribute. (12/14)
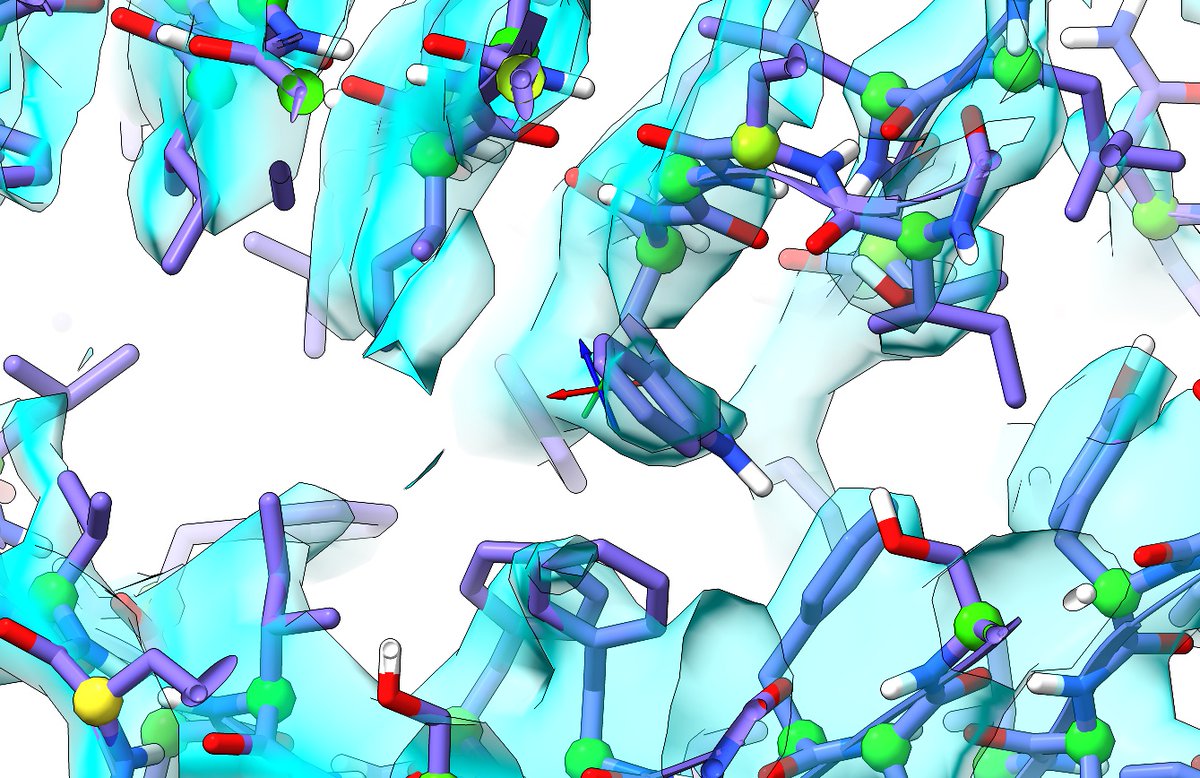
My part came relatively late in the game: specifically, rebuilding and refining an atomic model of the N439K RBD:ACE2 complex against a 2.8 Angstrom crystal dataset gathered by the @Vir_Biotech team; and helping build out the full N-linked glycans... (13/14)
... to provide a solid starting model for simulations by @jchodera and his team to explore the potential conformational plasticity of RBD:ACE2 variants outside the constraints of the crystallographic environment. (14/14)
• • •
Missing some Tweet in this thread? You can try to
force a refresh