A more extreme illustration of the problem: yes, this is a single residue (JSG, found in 6s8h and 6mhu). What it actually *is* is E. coli lipopolysaccharide (LPS) - how it appears in the database is {deep breath}:
https://twitter.com/CrollTristan/status/1357659731691773953
(2~{r},4~{r},5~{r},6~{r})-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-2-[(2~{r},4~{r},5~{r},6~{r})-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-5-[(2~{s},3~{s},4~{r},5~{r},6~{r})-6-[(1~{s})-1,2-bis(oxidanyl)ethyl]-4-[(2~{r},3~{s},4~{r},5~{s},6~{r})-6-[(1~{s})-2-[(2~{s},3~{s},4~{s},5~{s},6~{r})-...
...6-[(1~{s})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{r},3~{s},4~{r},5~{r},6~{r})-5-[[(3~{r})-3-dodecanoyloxytetradecanoyl]amino]-6-...
...[[(2~{r},3~{s},4~{r},5~{r},6~{r})-3-oxidanyl-5-[[(3~{r})-3-oxidanyltetradecanoyl]amino]-4-[(3~{r})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{r})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-...
...bis(oxidanyl)oxane-2-carboxylic acid
... phew!
The thing is, there isn't just *one* LPS (this is biology - is there ever just one of anything?). Lipopolysaccharides are a whole family - here's the Wikipedia image for the general form:
... phew!
The thing is, there isn't just *one* LPS (this is biology - is there ever just one of anything?). Lipopolysaccharides are a whole family - here's the Wikipedia image for the general form:

Are we going to define a whole new ligand for every new one of these beasts (or pieces thereof)? God, I hope not... but if we look more closely at JSG, we can identify 12 logical cut sites (ester, amide and glycosidic bonds) making 13 blocks. A big advantage of doing it that way:
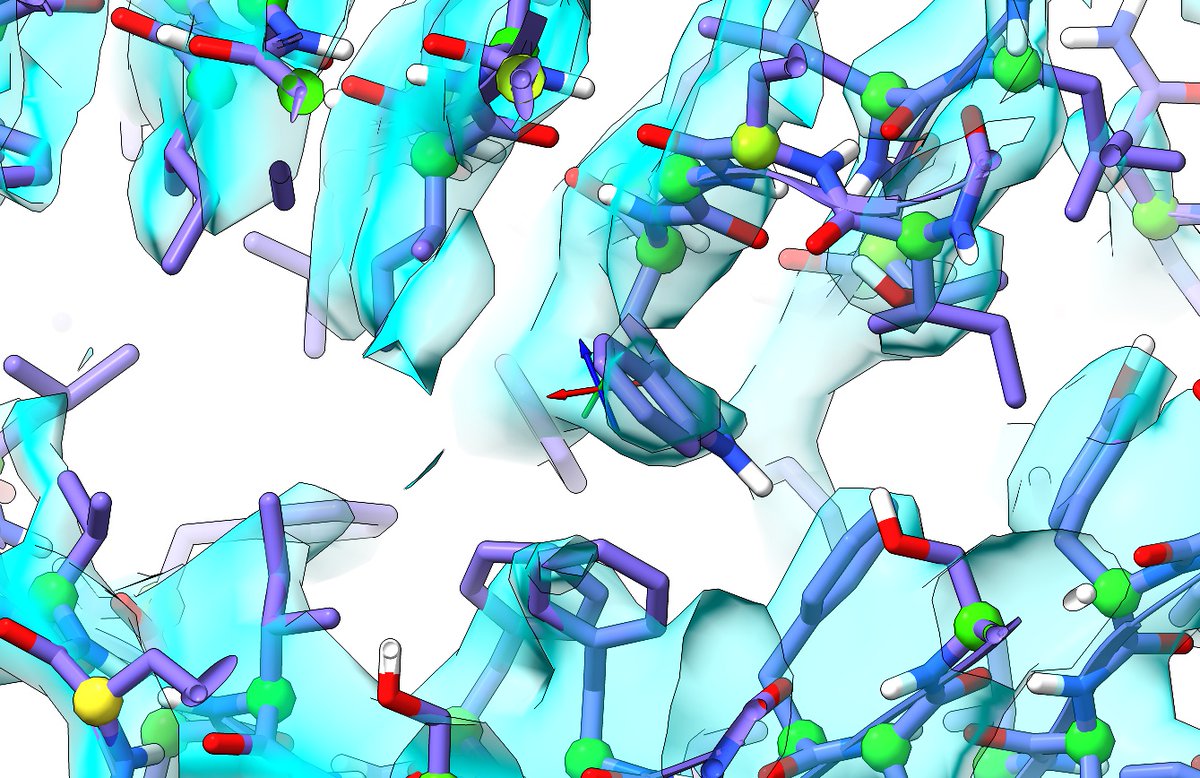
If we cut into individual residues like this, then it becomes easy for programs like @glycojones PRIVATEER to identify the sugars *as sugars*, which could have helped in this case - it would have flagged that 4 of the 6 rings are in unfavourable conformations.
• • •
Missing some Tweet in this thread? You can try to
force a refresh