Excited to present my first @capra_lab PhD project in today's @GeneticsSociety's @AJHGNews!!
Interested in #3Dgenome structure, complex trait #heritability, and/or evolutionary constraint?
cell.com/ajhg/fulltext/…
1/n
Interested in #3Dgenome structure, complex trait #heritability, and/or evolutionary constraint?
cell.com/ajhg/fulltext/…
1/n
By synthesizing topologically associating domain (TAD) maps across 37 diverse cell types with 41 genome-wide association studies (GWASs), we investigate the differences in disease association and evolutionary pressure on variation across the 3D genome landscape.
2/n
2/n

We know that TAD boundary disruption by SV can lead to developmental disease and cancers. (Thus, selection acts against these SVs).
But what about the relationship between common human variation in TAD boundaries (eg. SNPs) and associations with complex traits?
3/n
But what about the relationship between common human variation in TAD boundaries (eg. SNPs) and associations with complex traits?
3/n
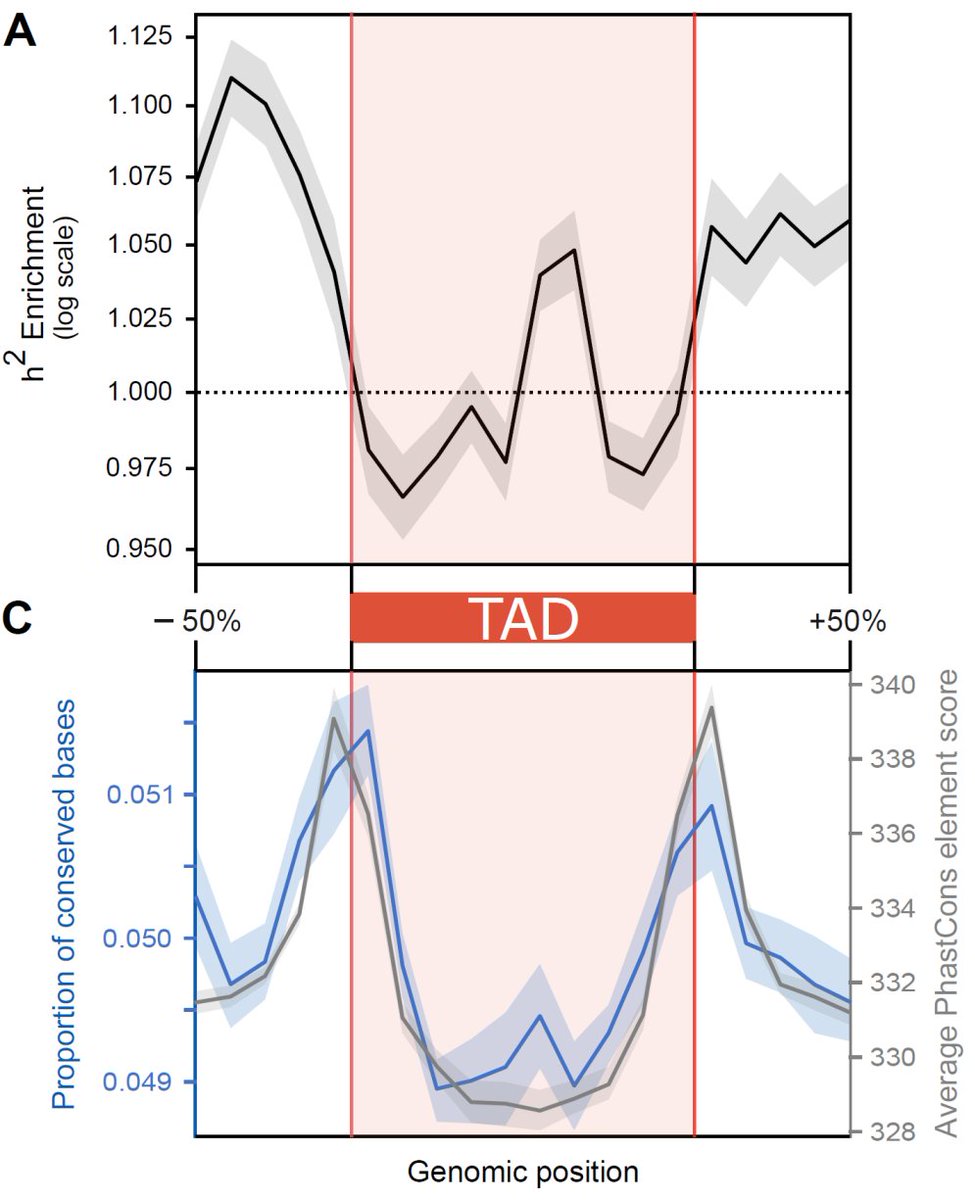
We use S-LDSC to estimate common-trait heritability (h2) enrichment across the 3D genome landscape and find that TAD boundaries (regions flanking TADs) are largely enriched for h2 and are more evolutionarily conserved at the base-pair level.
4/n
4/n
However, TAD boundaries are not all the same! We introduce a method for quantifying the stability of a TAD boundary across cell types and show that stable TAD boundaries are *further* enriched for trait h2 and evolutionary conservation (plus CTCF, housekeeping genes).
5/n
5/n

Finally, we hypothesized that variation in boundaries influences certain traits more. We show that hematologic/immunologic traits are the most enriched for trait h2 at TAD boundaries. Maybe the 3D genome plays differing roles in the genetic architecture of different traits?!
6/n
6/n

Please see the paper for more robust results, full figs, and discussion! We hope this provides another stepping stone towards understanding how specific genetic variants may affect 3D genome structure and disease risk in a cell-type-specific manner. Excited for the future!
7/n
7/n
This would not have been possible without great conversations/community and publically available data/methods. Specific thanks to:
@VUMCgenetics @VanderbiltMSTP @capra_lab
@yuefeng_1 lab 3D genome browser
@bmneale lab UKB GWAS
Price/Finucane Lab S-LDSC
@genome_gov funding
8/8
@VUMCgenetics @VanderbiltMSTP @capra_lab
@yuefeng_1 lab 3D genome browser
@bmneale lab UKB GWAS
Price/Finucane Lab S-LDSC
@genome_gov funding
8/8
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh