
Ct values can be used to estimate epidemic dynamics UPDATE! Ct values are expected to change depending on whether the epidemic is growing or declining, and we harness this to estimate the epidemic trajectory. Lots of cool new analyses and methods! 1/12
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…
Highlights:
- Cts from symptom-based surveillance change over time, but the effect is weaker
- Methods to infer incidence using single cross-sections of Cts
- Unbiased by changing testing coverage
- Gaussian process (wiggly line) model for incidence tracking using Ct values
2/12
- Cts from symptom-based surveillance change over time, but the effect is weaker
- Methods to infer incidence using single cross-sections of Cts
- Unbiased by changing testing coverage
- Gaussian process (wiggly line) model for incidence tracking using Ct values
2/12
This work is *JOINTLY led* with @LeekShaffer and PI’d by @michaelmina_lab. Thank you also to the ever insightful @mlipsitch and to coauthors @SanjatKanjilal @gabriel_stacey and @nialljlennon. 3/12
Premise: times since infection depend on the epidemic trajectory. Distributions of randomly sampled viral loads proxy times since infection. With calibration, Ct values can estimate growth rate. We focus on qPCR in SARS-CoV-2, but the principle applies to any outbreak. 4/12
Result 1: viral loads are shifted higher (Cts lower) during epidemic growth and lower (Cts higher) during decline when individuals are sampled *based on the onset of symptoms*. We simulated linelist data under symptom-based surveillance and looked at TSI and Cts over time. 5/12 

This is crucial when considering virulence in emerging SARS-CoV-2 variants. Lower Cts over time do not *necessarily* mean newly dominant variants have higher virulence. If incidence of a new variant is increasing, then we expect to see more recent infections and lower Cts. 6/12
**However, the effect is smaller than under random surveillance, so I would not rule out the possibility of increased virulence.** But important to consider. Thank you to @charliewhittak for chatting through this! 7/12
Result 2: we reconstructed the epidemic curve using single-cross sectional samples from well-observed nursing homes, finding that single cross sections using the full Ct distribution provided similar insights to point prevalence across three sample times. 8/12 
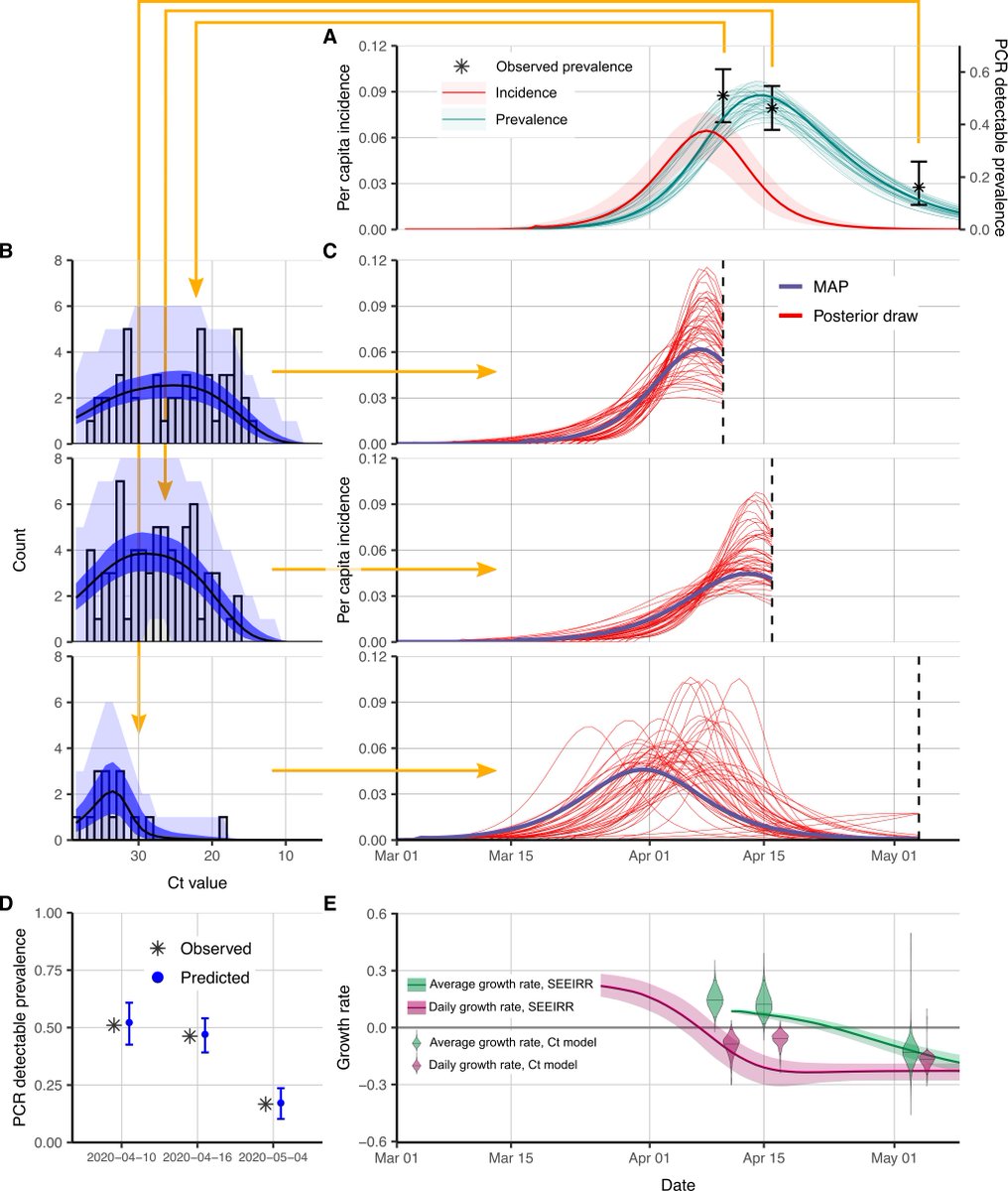
Result 3: we compared Ct-based to case-count based methods when testing is changing. Rt estimates are biased when testing is increasing or decreasing (not a problem with the method, just the data!). Our method uses the Ct distribution so does not care about test numbers. 9/12 

Result 4: we use multiple cross-sectional samples to reconstruct incidence without making assumptions about the trajectory shape (a Gaussian “wiggly” process model). We can track the incidence curve in MA using routinely collected hospital tests. 10/12 

… and here is a gif that reminds me of a nematode worm. Every week we add on a new cross section of Cts and accurately track true incidence (in simulation, red line). 11/12
Conclusion: we are generating loads of (semi) quantitative data in the form of Cts. We can harness these to get unbiased estimates of the epidemic trajectory. Hopefully these ideas will help public health surveillance efforts and interpret data in the light of new variants. 12/12
Individuals tested due to recent symptom onset are more likely to have been recently infected with a short incubation period during epidemic growth than during epidemic decline, where more onsets are from older infections with longer incubation periods.
journals.plos.org/ploscompbiol/a…
journals.plos.org/ploscompbiol/a…
Original thread here:
https://twitter.com/jameshay218/status/1316202275048579072
• • •
Missing some Tweet in this thread? You can try to
force a refresh


