Update from @my_helix SARS-CoV-2 surveillance program
Check public.tableau.com/profile/helix6…
🧵 with FL, TX, GA, CA & MA + results on identification of B.1.351 & P.1 !!
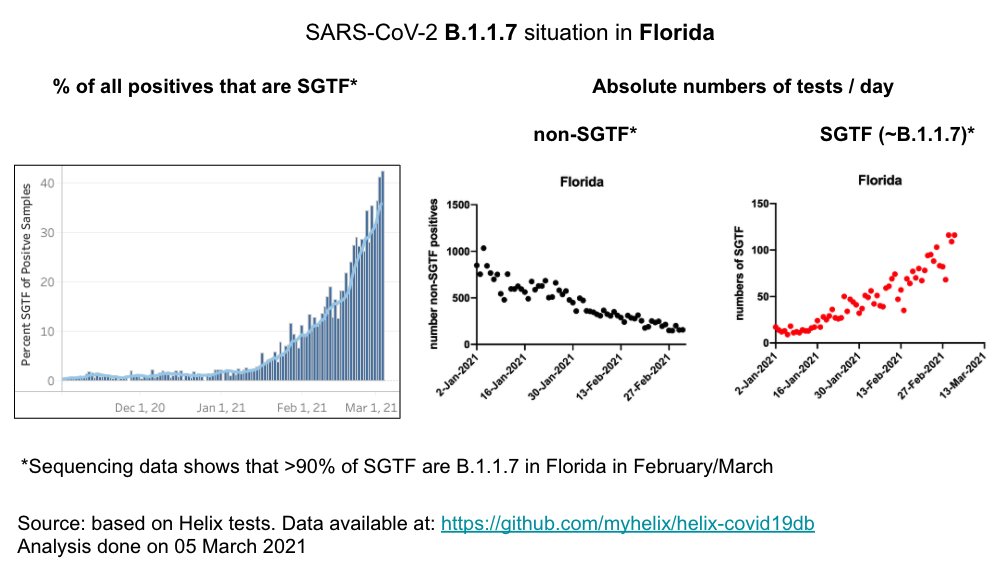
FL: B.1.1.7 is ~40% of positives. 📈 about as predicted
Absolute numbers of B117 up despite overall 📉 in cases
Check public.tableau.com/profile/helix6…
🧵 with FL, TX, GA, CA & MA + results on identification of B.1.351 & P.1 !!
FL: B.1.1.7 is ~40% of positives. 📈 about as predicted
Absolute numbers of B117 up despite overall 📉 in cases
2/
Texas
March 3: B117 were ~30% of positive tests.
26% with 5-day avg.
Absolute numbers of SGTF (a great proxy for B117 now) also going up despite big decrease in overall cases (as seen overall in US)
Note: this is only based on Helix data
Texas
March 3: B117 were ~30% of positive tests.
26% with 5-day avg.
Absolute numbers of SGTF (a great proxy for B117 now) also going up despite big decrease in overall cases (as seen overall in US)
Note: this is only based on Helix data

3/
Georgia
March 3: B117 represents ~26% of positive tests.
Absolute numbers of SGTF (a great proxy for B117 now) also going up despite big decrease in overall cases (as seen overall in US)
Georgia
March 3: B117 represents ~26% of positive tests.
Absolute numbers of SGTF (a great proxy for B117 now) also going up despite big decrease in overall cases (as seen overall in US)

4/
California
March 4: B117 is 27% of positives but 5-day avg is lower at 18%.
Absolute numbers of B117 do go up despite the big decrease in number of total cases, but it is perhaps less striking than FL, GA or TX.
California
March 4: B117 is 27% of positives but 5-day avg is lower at 18%.
Absolute numbers of B117 do go up despite the big decrease in number of total cases, but it is perhaps less striking than FL, GA or TX.

5/
Massachusetts
MA is so interesting because in Nov & Dec it had a STABLE 3-5% of tests that were SGTF (S gene target failure). These were NOT B.1.1.7 (they were B.1.375 and other lineages with H69del).
As soon as B117 was detected in early Feb, fraction of SGTF started 📈
Massachusetts
MA is so interesting because in Nov & Dec it had a STABLE 3-5% of tests that were SGTF (S gene target failure). These were NOT B.1.1.7 (they were B.1.375 and other lineages with H69del).
As soon as B117 was detected in early Feb, fraction of SGTF started 📈

6/
Info on other variants
B.1.351 : variant of concern first identified in South Africa
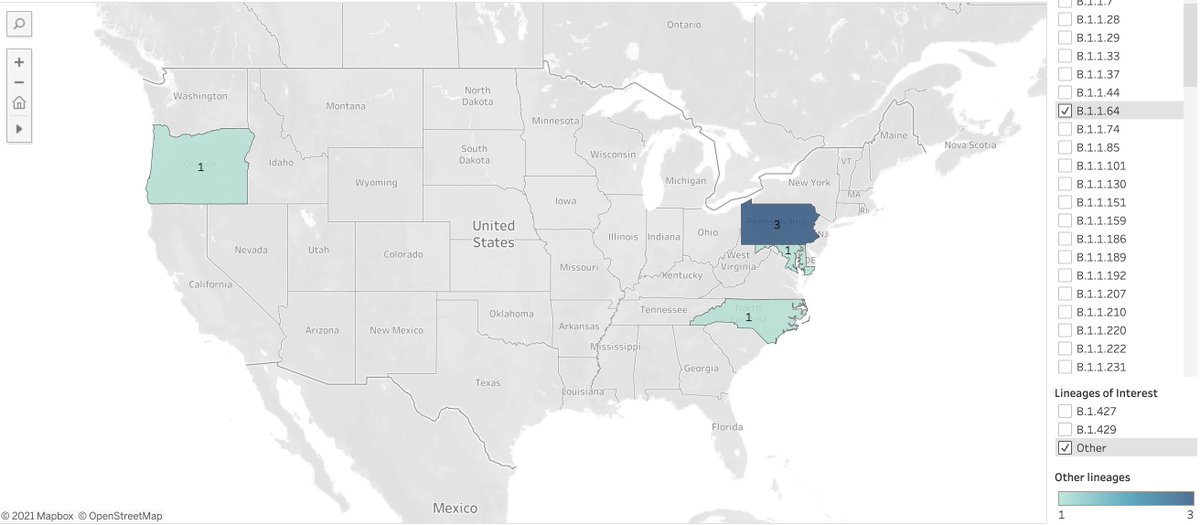
4 cases: 1 in CT, 1 in FL, 1 in GA, 1 in NC
P.1 : variant of concern first identified in Brazil
3 cases: all in FL
All the data: github.com/myhelix/helix-… (thanks @simonjwhite71 @Maglaar)
Info on other variants
B.1.351 : variant of concern first identified in South Africa
4 cases: 1 in CT, 1 in FL, 1 in GA, 1 in NC
P.1 : variant of concern first identified in Brazil
3 cases: all in FL
All the data: github.com/myhelix/helix-… (thanks @simonjwhite71 @Maglaar)

7/
Other vars NOT of concern
B.1.525: 25 times incl. 10 in GA
Defining mutations include:
S:E484K
S:H69delV70del -> it is a SGTF (keep in mind)
Orf1a:9bp del also seen in B.1.1.7, B1.351 & P.1
Is it of interest? Do we know if the 9bp del in Orf1a has impact?
@firefoxx66
Other vars NOT of concern
B.1.525: 25 times incl. 10 in GA
Defining mutations include:
S:E484K
S:H69delV70del -> it is a SGTF (keep in mind)
Orf1a:9bp del also seen in B.1.1.7, B1.351 & P.1
Is it of interest? Do we know if the 9bp del in Orf1a has impact?
@firefoxx66

8/
Source for screenshot ☝️ cov-lineages.org/global_report_…
Curious to know who decides (& how) whether a variant is of interest.
& is there a resource gathering functional info on mutations outside Spike protein?
Orf1a 9bp del?
N:M1X?
etc
do they matter?
@K_G_Andersen @trvrb
Source for screenshot ☝️ cov-lineages.org/global_report_…
Curious to know who decides (& how) whether a variant is of interest.
& is there a resource gathering functional info on mutations outside Spike protein?
Orf1a 9bp del?
N:M1X?
etc
do they matter?
@K_G_Andersen @trvrb
• • •
Missing some Tweet in this thread? You can try to
force a refresh