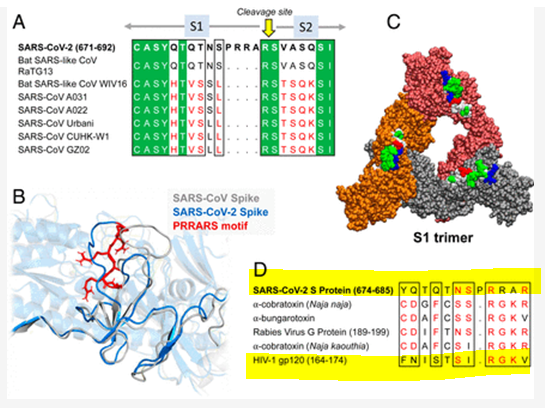
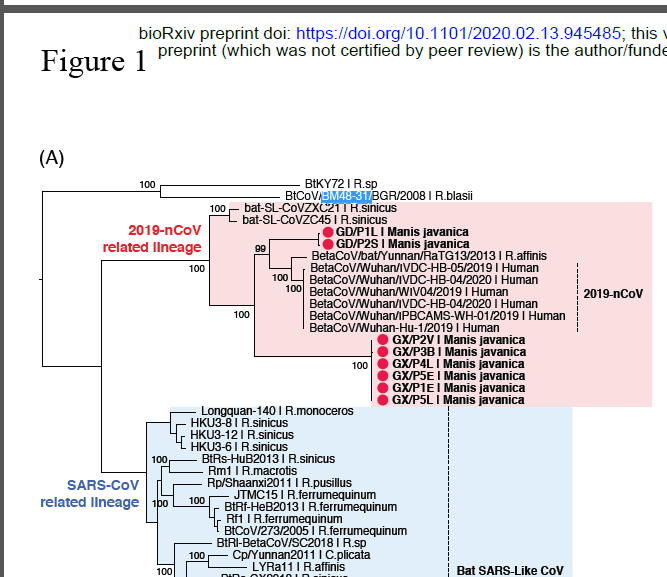
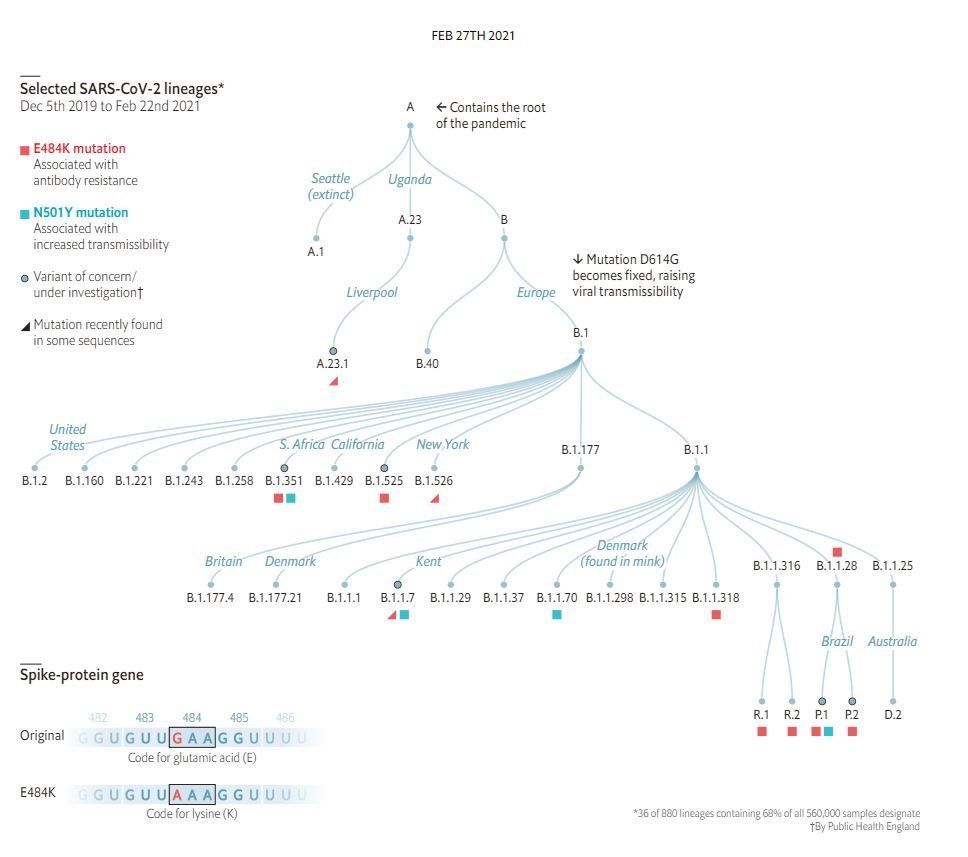
Compilation Thread on ORF8 ORF8 or Fate?
1. ZLS (2018):
"Considering the variability of orf8 in bats, civets and humans, investigating the function of orf8 is a priority, particularly the contribution of these different variants to viral pathogenesis."
1. ZLS (2018):
"Considering the variability of orf8 in bats, civets and humans, investigating the function of orf8 is a priority, particularly the contribution of these different variants to viral pathogenesis."
https://twitter.com/BillyBostickson/status/1251750041460465665
2. ORF8 or Fate?
"In addition to receptor binding, proteolytic cleavage of S & potentially other mutations that affect virion & trimer stability may be important for virus transmissibility in different hosts, & these factors need to be studied further."
nature.com/articles/s4157…
"In addition to receptor binding, proteolytic cleavage of S & potentially other mutations that affect virion & trimer stability may be important for virus transmissibility in different hosts, & these factors need to be studied further."
nature.com/articles/s4157…
3. ORF8 or Fate?
Overwhelmingly these viruses had mutations or
29 deletions in ORF8 - associated with reduced replicative fitness of the virus.. may be associated with host adaptation
via @CarltheChippy
382-nt deletion during early evolution of SARS-CoV
biorxiv.org/content/10.110…
Overwhelmingly these viruses had mutations or
29 deletions in ORF8 - associated with reduced replicative fitness of the virus.. may be associated with host adaptation
via @CarltheChippy
382-nt deletion during early evolution of SARS-CoV
biorxiv.org/content/10.110…
4. ORF8 or Fate?
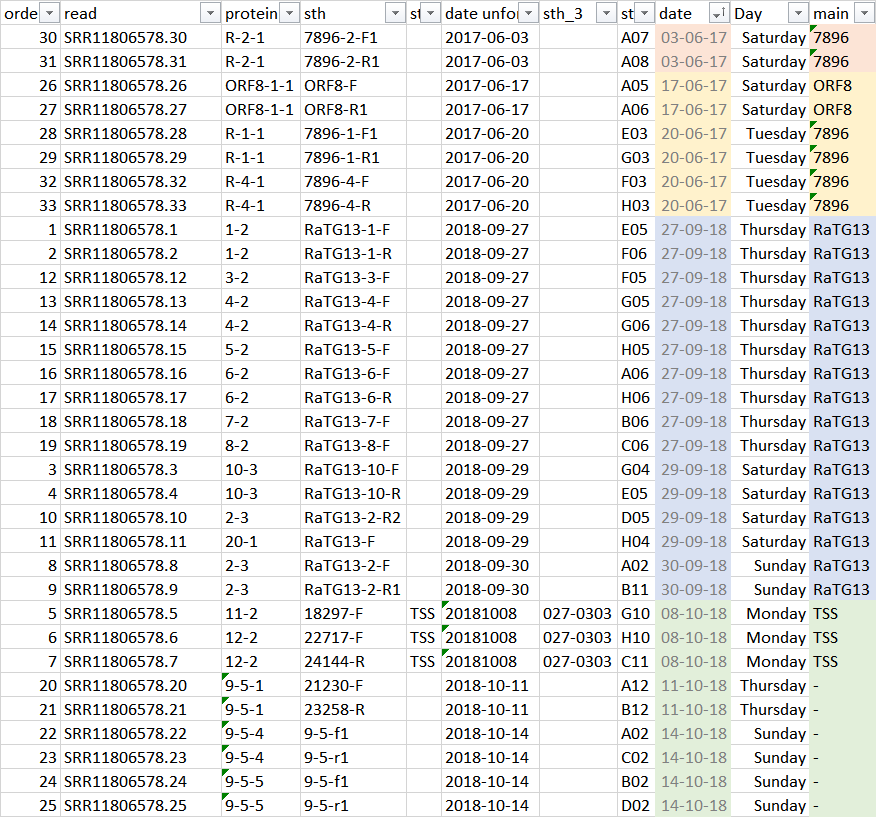
@franciscodeasis reviewed the list of read filenames according to dates and labels:
- Sat 2017-06-03 (7896)
- Sat 2017-06-17 to Tue 2017-06-20 (7896 and ORF8)
- Thu 2018-09-27 to Sun 2018-09-30 (RaTG13)
- Mon 2018-10-08 to Sun 2018-10-14 (no label and TSS)
@franciscodeasis reviewed the list of read filenames according to dates and labels:
- Sat 2017-06-03 (7896)
- Sat 2017-06-17 to Tue 2017-06-20 (7896 and ORF8)
- Thu 2018-09-27 to Sun 2018-09-30 (RaTG13)
- Mon 2018-10-08 to Sun 2018-10-14 (no label and TSS)
5. ORF8 or Fate?
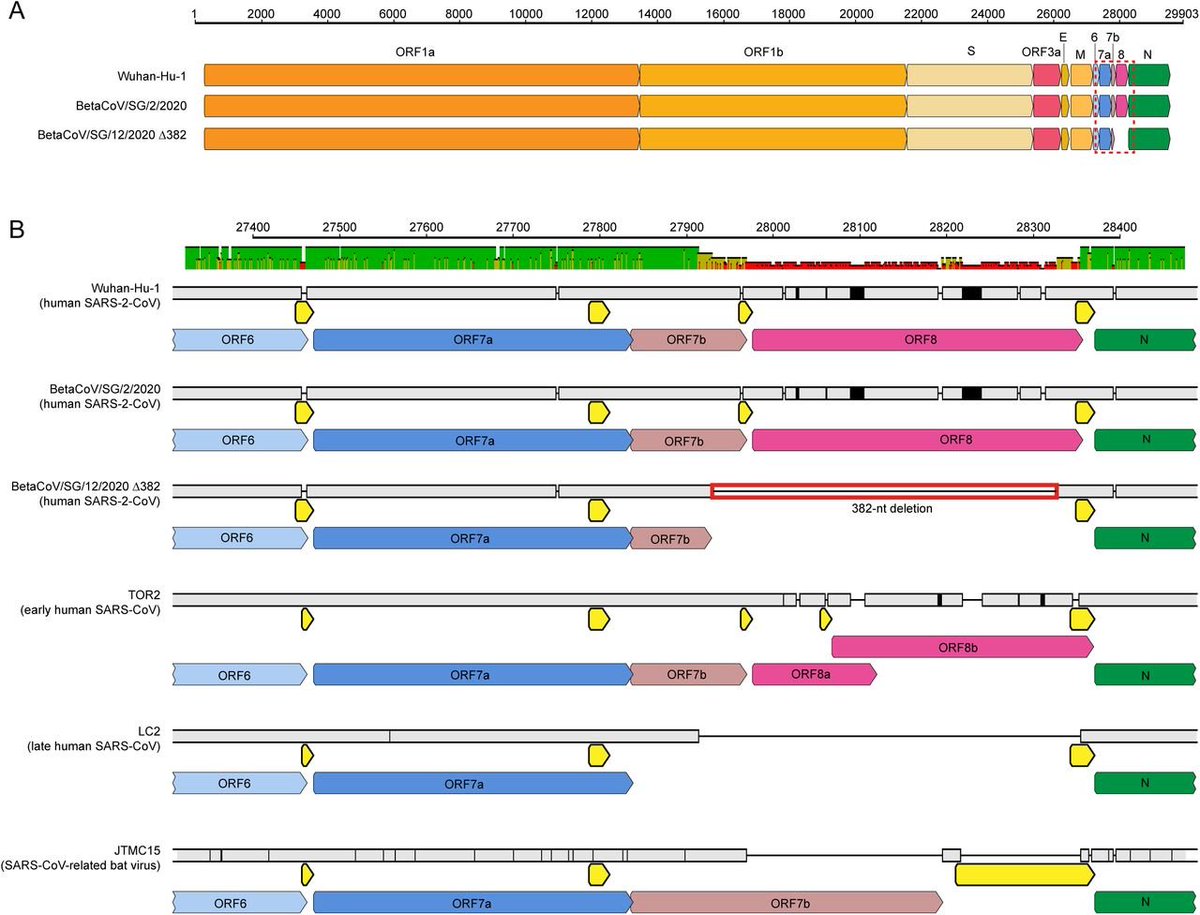
What is ORF8? This is Orf8!

What is ORF8? This is Orf8!
https://twitter.com/BillyBostickson/status/1283835714585542656
6. ORF8 or Fate? keep on saying it!
orf8 NSP12 deletion found in India
biorxiv.org/content/10.110…

orf8 NSP12 deletion found in India
biorxiv.org/content/10.110…
https://twitter.com/BillyBostickson/status/1283842341535535105

7. ORF8 or Fate?
Via @ConsciousEvolu1
What is critically important is that Orf8 and surface Glycoproteins create Porphyrin penetration of the Cell. Resulting in more Severe Infections, Bio-chemic Processes interuption, Heme cleavage and Hypoxia.
researchgate.net/publication/33…
Via @ConsciousEvolu1
What is critically important is that Orf8 and surface Glycoproteins create Porphyrin penetration of the Cell. Resulting in more Severe Infections, Bio-chemic Processes interuption, Heme cleavage and Hypoxia.
researchgate.net/publication/33…
8. ORF8 or Fate?
via @Rossana38510044
"The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I"
biorxiv.org/content/10.110…
via @Rossana38510044
"The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I"
biorxiv.org/content/10.110…
10. ORF8 or Fate? Stress Trigger!
"However, 2 proteins (orf8 & orf10) in SARS-CoV-2 have no homologous proteins in SARS-CoV. The amino acid sequence of orf8 in SARS-CoV-2 is different from sequences of conserved orf8 or orf8b derived from human SARS-CoV"
mdpi.com/1999-4915/12/2…
"However, 2 proteins (orf8 & orf10) in SARS-CoV-2 have no homologous proteins in SARS-CoV. The amino acid sequence of orf8 in SARS-CoV-2 is different from sequences of conserved orf8 or orf8b derived from human SARS-CoV"
mdpi.com/1999-4915/12/2…

11.ORF8 or Fate! Coincidence? No, Trust a Me, it's Natural!
1. Orf8 "combined" with Orf10 to create a Porphyrin Heme Cleave and Orf8 Porphyrin Infectivity
2. WIV were already studying the RaTG13 and 7896 Clade Spike Proteins and Orf8 back in 2018
,
1. Orf8 "combined" with Orf10 to create a Porphyrin Heme Cleave and Orf8 Porphyrin Infectivity
2. WIV were already studying the RaTG13 and 7896 Clade Spike Proteins and Orf8 back in 2018
,

12. via @CZilcho
not a single live virus. genome looks like the version of a "virus" that has been compiled from known RaTG13 contigs & consensus seq from RmYN02 (ORF 1ab, 3a, E, M, 6, 7a) + some unknown seq (passaged viruses?) for S, & 3' from ORF8, N, using Shiver or similar.
not a single live virus. genome looks like the version of a "virus" that has been compiled from known RaTG13 contigs & consensus seq from RmYN02 (ORF 1ab, 3a, E, M, 6, 7a) + some unknown seq (passaged viruses?) for S, & 3' from ORF8, N, using Shiver or similar.

13. ORF8 or Fate? ORF8 similarities
Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus
Ben Hu (2017)
ncbi.nlm.nih.gov/pmc/articles/P…
Peppered with 91 references to Orf8



Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus
Ben Hu (2017)
ncbi.nlm.nih.gov/pmc/articles/P…
Peppered with 91 references to Orf8




15. ORF8 or Fate?
1. ORF8 enhances the viral replication.
2. enhanced ORF8s are main factors contributing to transmission, virulence and host adaptability of CoVs.
3. enhanced ORF8s increase the efficiencies in viral entry into cells and replication
biorxiv.org/content/10.110…
1. ORF8 enhances the viral replication.
2. enhanced ORF8s are main factors contributing to transmission, virulence and host adaptability of CoVs.
3. enhanced ORF8s increase the efficiencies in viral entry into cells and replication
biorxiv.org/content/10.110…

16. ORF8 or Fate?
Back in Time to an ORF8 rich bat cave with WIV and @amicocolorido
Distribution of SARSr-CoVs highly similar to SARS-CoV in the variable S, ORF3 and ORF8 genes in the single cave.
ncbi.nlm.nih.gov/pmc/articles/P…

Back in Time to an ORF8 rich bat cave with WIV and @amicocolorido
Distribution of SARSr-CoVs highly similar to SARS-CoV in the variable S, ORF3 and ORF8 genes in the single cave.
ncbi.nlm.nih.gov/pmc/articles/P…


17. Some naughty but nice ORF8y images collated by @BidoliNicola
https://twitter.com/BidoliNicola/status/1346784101060272129
18. ORF8 or Fate? Back to SARS
ncbi.nlm.nih.gov/pmc/articles/P…
Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination
ncbi.nlm.nih.gov/pmc/articles/P…
Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination
https://twitter.com/BillyBostickson/status/1350024332072796164
19. ORF8 or Fate?
Latest Findings - ORF8's secrets exposed
" two unique regions: one that is only present in SARS-CoV-2 and its immediate bat ancestor, and one that is absent from any other coronavirus,"
medicalxpress.com/news/2021-03-g…

Latest Findings - ORF8's secrets exposed
" two unique regions: one that is only present in SARS-CoV-2 and its immediate bat ancestor, and one that is absent from any other coronavirus,"
medicalxpress.com/news/2021-03-g…


20. ORF8 or Fate? WTF is this?
Paper referenced in above article:
The odd structure of ORF8: Mapping the coronavirus protein linked to disease severity
phys.org/news/2021-01-o…


Paper referenced in above article:
The odd structure of ORF8: Mapping the coronavirus protein linked to disease severity
phys.org/news/2021-01-o…



21. ORF8 or Fate? via @Harvard2H
Divergence between Orf7a & Orf8 is exceptionally idiosyncratic, as Orf7a is more constrained, whereas Orf8 is subject to rampant change, a peculiar feature that may be related to hitherto unknown viral infection strategies
mbio.asm.org/content/mbio/1…


Divergence between Orf7a & Orf8 is exceptionally idiosyncratic, as Orf7a is more constrained, whereas Orf8 is subject to rampant change, a peculiar feature that may be related to hitherto unknown viral infection strategies
mbio.asm.org/content/mbio/1…



22. "Barring a Technical Artefact"
"Protein interactions of SARS-CoV-2 exhibit such a sharp contrast between Orf7a and Orf8 that, barring a technical artifact with respect to coverage..Orf7a generate variants, such as Orf8, wreaking havoc through immune evasion in the host cell"
"Protein interactions of SARS-CoV-2 exhibit such a sharp contrast between Orf7a and Orf8 that, barring a technical artifact with respect to coverage..Orf7a generate variants, such as Orf8, wreaking havoc through immune evasion in the host cell"

23. ORF8 or Fate? via @Undergroundsar3
ORF8 is involved somehow in apoptosis/viral cytolysis pathway and functions to optimize spread of virons while cloaking the infected host cell from CD8+ killer T-cell surveillance.
ORF8 is involved somehow in apoptosis/viral cytolysis pathway and functions to optimize spread of virons while cloaking the infected host cell from CD8+ killer T-cell surveillance.
24. ORF8 or Fate? @SBgrid
1. Solved structure of SARS-CoV-2 ORF8 uncovers vital role in immune suppression & evasion
pnas.org/content/118/2/…
2. Covalent dimer is an "evolutionarily" recent addition unique to SARS-CoV-2
3. SARS-CoV-2 ORF8 contains a large, unstructured insertion

1. Solved structure of SARS-CoV-2 ORF8 uncovers vital role in immune suppression & evasion
pnas.org/content/118/2/…
2. Covalent dimer is an "evolutionarily" recent addition unique to SARS-CoV-2
3. SARS-CoV-2 ORF8 contains a large, unstructured insertion


25. ORF8 or Fate?
Question for Hans Kristian from @Undergroundsar3
What are your thoughts on the CD8+ t-cell story in the milieu of the host (vs. test tube humoral only neutralization assays) where ORF8 is expressed?
Referenicing:
immunology.sciencemag.org/content/6/57/e…
Question for Hans Kristian from @Undergroundsar3
What are your thoughts on the CD8+ t-cell story in the milieu of the host (vs. test tube humoral only neutralization assays) where ORF8 is expressed?
https://twitter.com/Undergroundsar3/status/1367596736101945352
Referenicing:
immunology.sciencemag.org/content/6/57/e…
27. ORF8 or Fate?
Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene
researchgate.net/publication/34…


Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene
researchgate.net/publication/34…



28. ORF8 or Fate?
Characterization of accessory genes in coronavirus genomes
link.springer.com/article/10.118…

Characterization of accessory genes in coronavirus genomes
link.springer.com/article/10.118…


29. ORF8 or Fate? To be or not to be?
SARS-CoV-2 variants combining spike mutations & absence of ORF8 may be more transmissible & require close monitoring
researchgate.net/publication/34…
SARS-CoV-2 phylogeny highlighting ORF8-deficient variants resulting from nonsense mutation E64stop.
SARS-CoV-2 variants combining spike mutations & absence of ORF8 may be more transmissible & require close monitoring
researchgate.net/publication/34…
SARS-CoV-2 phylogeny highlighting ORF8-deficient variants resulting from nonsense mutation E64stop.

30. From 2018 to 2021 with ORF8...or Fate?
unroll @threadreaderapp
save @buzz_chronicles
unroll @threadreaderapp
save @buzz_chronicles

• • •
Missing some Tweet in this thread? You can try to
force a refresh