
Coming up in a few minutes, Clive Brown @the_taybor, with an update entitled “Nobody Expects the Strandish Exposition”. Follow the talk live in this thread #nanoporeconf
CB: Our goal is to enable the analysis of anything by anyone, anywhere. Here are some examples of global use: nanoporetech.com/portable-seque…
CB: As a reminder, this is how nanopore sequencing works. #nanoporeconf
CB: @nanopore has an information-rich signal which can be deconvoluted in real time. Basecalling updates have driven considerable improvements over the past two years. 

We have moved onto self supervised methods of data analysis, and have bulked out our machine learning and data analysis teams #nanoporeconf
CB: the Bonito basecaller, introduced as a research basecaller in late 2020, is now in MinKNOW is reaching modal single read accuracy of 98.3% #nanoporeconf
https://twitter.com/kirk3gaard/status/1328087574951432192?lang=en-gb
CB: The Q20+ chemistry is delivering raw read accuracies of 99%+ in developers hands
https://twitter.com/AlbertVilella/status/1384846947446624257#nanoporeconf
CB: Q20+ chemistry is now moving from developers into early access – register your interest here register.nanoporetech.com/q20plus-chemis…
CB: the Q20+ kit is compatible with MinION and GridION now #nanoporeconf 

CB: we like to come back again and again to ideas and changing them to make them work. introducing Duplex #nanoporeconf
CB: new method, “Duplex”: following on. Enables nanopore devices to sequence a template and complement strand of a single molecule of DNA, in succession, in order to achieve very high accuracy sequencing results. #nanoporeconf
CB: terminology: Simplex= single strand. Both strands = duplex #nanoporeconf
CB: goal of Duplex experiments is to achieve a high proportion of instances where the complement follows the template strand through the nanopore and gives the system ‘two looks at the same sequence pairs’. #nanoporeconf
CB: Current runs are routinely achieving 40% of data from Duplex pairs, with 50% the current highest and 75% is targeted after further optimisation. #nanoporeconf 

CB: upgraded enzyme to E8.1, improvement the movement quality on the complement strand. Complement quality is almost the same as the template strand #nanoporeconf
CB: Duplex is compatible with the ultra-long kit capable of N50s greater than 100kb. Further optimisation required. Longest duplex pair to date is 442kb
#nanoporeconf
#nanoporeconf
CB: Duplex can currently achieve modal accuracy of ~Q29 for a single double stranded molecule of DNA #nanoporeconf 
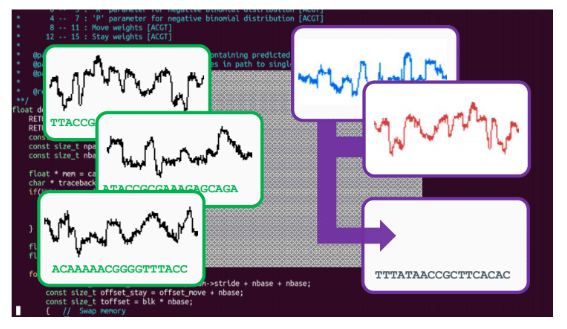
CB: the intrinsic data quality on this platform is extremely high, and we are now achieving that because of the software #nanoporeconf
CB: a key differentiator is the length of the duplex reads. The longest Duplex molecule sequenced to Q30 to date is 156kb. #nanoporeconf
CB: we can't see any substantial difference between our Duplex reads and another leading platform's high accuracy mode #nanoporeconf
CB: revision. Without a motor, single stranded DNA translocates the pore very quickly ~thousands of bases per second and thought to be “slip-stick” hard to control
― (blunt double stranded DNA won’t strip through) #nanoporeconf
― (blunt double stranded DNA won’t strip through) #nanoporeconf
CB: controlled movement is needed through the pore, for high accuracy sequencing. => motors #nanoporeconf
CB: Current nanopore chemistries sequence DNA as it is driven from the ‘top’ of the flow cell through the nanopore to the area underneath; this is known as ‘Inny’. #nanoporeconf 

CB: many potential benefits of deploying the opposite ‘Outy’ direction, one format being where DNA threads from the 3’ end first, still moving 5’ to 3’. different DNA orientation may produce a different error profile #nanoporeconf
CB: we decided to start looking at 'outy' again. Here's an Outy mode: #nanoporeconf 

CB: …potential to enable approximate fragment sizing before sequencing – and therefore enable pre-selection of the desired read lengths. The method can also be used to perform controlled re-reading of native strands. #nanoporeconf
CB: ‘Adaptive accuracy’ possible with ‘outy’ mode #nanoporeconf 

CB: we can re-read a fragment on an indefinite loop. The signal from the first read is the same as the 4th/5th.. you can basecall as you go, to a point where you are happy with the accuracy and then you move on to the next fragment #nanoporeconf
CB: a reminder of Adaptive Sampling – real-time acceptance or rejection of molecules from a nanopore based on sequence or signal – to enrich/deplete the sample for specific targets of interest
nanoporetech.com/about-us/news/… #nanoporeconf
nanoporetech.com/about-us/news/… #nanoporeconf
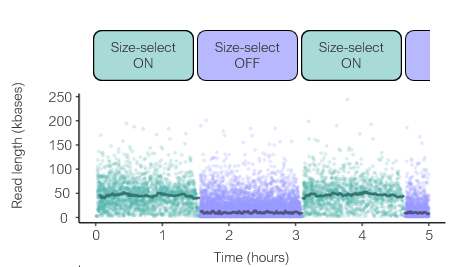
CB: could size the fragment before sequencing it using "inny", with current Q20 chemistry #nanoporeconf 

CB: this is what that looks like #nanoporeconf 
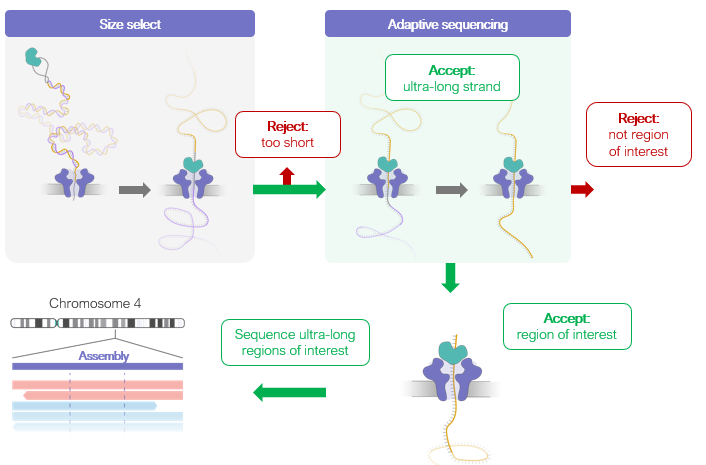
CB: could combine Adaptive sampling & ultra-long sequencing & Q20+. “Adaptive finishing” for faster & even coverage and finishing, by proactive selection of ultra-long fragments that covered regions of low, or no coverage. #nanoporeconf 
CB: if the input fragment size distribution is long enough, you should be able to adaptively select length of sample molecules, and adaptively sample haplotypes. I believe any genome can be assembled and closed with nanopore only.#nanoporeconf
CB: Next Generation Flongle. Developing new flow cells for the Flongle adapter, based on plastic not silicon for low cost. Plastic & silver chloride => aim for better signal/noise, cheaper/scalable manufacturing and stable currents for long run times. #nanoporeconf
CB: part of getting rid of the lab is getting rid of the pipette, and we want to do that with Flongle. We've been developing companion sample preps - should just need a swab. Prep too early to show today but big ambition #nanoporeconf
CB: CB: Custom ASIC collects data from individual nanopore channels; Low power ASICs in development to support pipeline eg SmidgION, Plongle 

CB: there is no point having a SmidgION mobile phone sequencer, if you need a bunch of lab kit. We're working on it #nanoporetech
CB: PromethION 48 generating very high outputs: 10Tb run record recently. Composed of 48 flow cells so modular design…. Users are scaling up with PromethION to very high throughput facilities with automation #nanoporeconf
CB: we have dramatically improved the electronics inside the box for PromethION. Now. Here's a new product #nanoporeconf
CB: we are developing PromethION 2. P2 will run up to two PromethION flow cells at once, includes compute. Starter pack $60k includes 48 flow cells. #nanoporeconf 

CB: pricing is provisional! But will be competitive #nanoporeconf
CB: why do P2? Two PromethION flow cells in small format keeps nanopore a step ahead of other devices in this range. P2 would be ideal for studying large genomes/ transcriptomes, easy with automation. You can register your interest here register.nanoporetech.com/p2-device
CB: P2 solo: Without integrated compute, to connect two flow cells to your own computer or GridION #nanoporeconf 

CB: if you make the investment in @nanopore, we will keep you ahead by upgrading your technology with all the latest improvements #nanoporetech
CB: a summary of the latest performance stats - for full details nanoporetech.com/accuracy
CB: We are doing a lot of work on the idea for writing DNA, and protein sequencing. We will say more later #nanoporeconf
CB: We have started a pilot programme called org.one, where we support free flow cells for sequencing of critically endangered species, to help conservationists get to grips with the current biodiversity crisis org.one/oo #nanoporeconf 

CB: we would like to support a ground-up movement to enable local sequencing of these critically endangered species, rather than conservationists sending their samples to wealthier countries for sequencing #nanoporeconf org.one/oo
CB: And that's the end of my update. #nanoporeconf
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh




