As promised, here is my #tweetorial for our @capra_lab @NatureComms paper! Interested in complex trait #genetics, #heritability, #Evolution , or #Neanderthals ? 🧬
Paper: nature.com/articles/s4146…
"Behind the paper" article: natureecoevocommunity.nature.com/posts/uncoveri…
1/
Paper: nature.com/articles/s4146…
"Behind the paper" article: natureecoevocommunity.nature.com/posts/uncoveri…
1/
We have examples of how Neanderthals gave Eurasians individual genetic variants that contribute to traits; however, most medically/evolutionarily relevant traits are complex, with contributions from thousands of parts of the genome. We wanted something more comprehensive...
2/
2/
We use partitioned heritability to investigate the relationship between introgression and diverse traits. First, we show that genomic regions with Neanderthal ancestry are depleted of heritability for all traits considered, except those related to skin and hair.
3/
3/

This expands upon previous findings showing that regions with Neanderthal ancestry are depleted for background selection and functional annotations. Not only are these regions depleted for functional annotations, but they are depleted for association with most human traits!
4/
4/
Then, for the introgressed variants specifically, we ask: how do they contribute to the genetics/heritability of 400+ traits? (thanks @uk_biobank!)
*Enriched for contribution to skin/bone/WBC/respiratory traits
*Depleted for contribution to cognition/procreation
5/
*Enriched for contribution to skin/bone/WBC/respiratory traits
*Depleted for contribution to cognition/procreation
5/

Partitioned heritability quantifies the overall contribution to variation in traits, but does not consider the direction of effect.
(Think: Do Neanderthal variants contribute to height? VERSUS Are Neanderthal variants more likely to make me taller or shorter?)
6/
(Think: Do Neanderthal variants contribute to height? VERSUS Are Neanderthal variants more likely to make me taller or shorter?)
6/
We used two approaches to test for directionality but, overall:
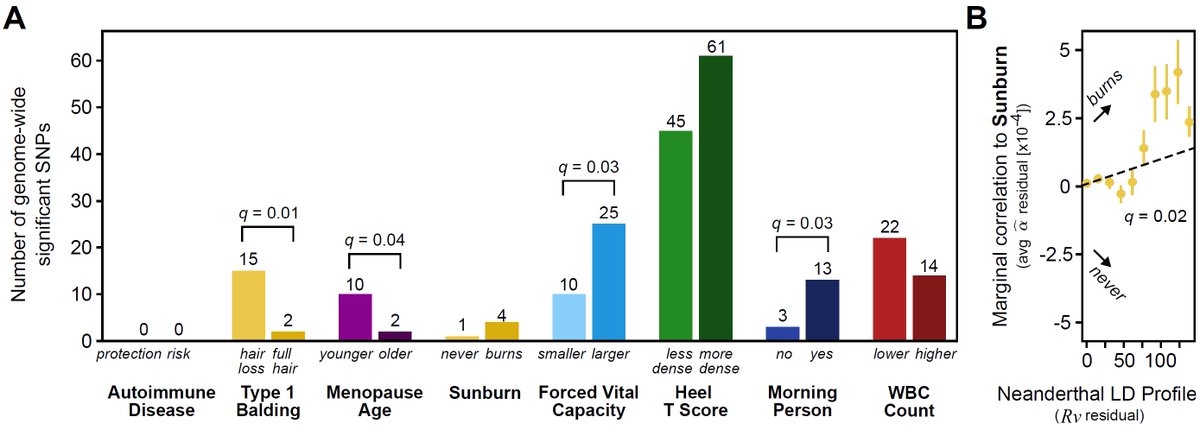
For some traits, Neanderthal variants contribute a directional effect (balding risk, younger menopause, larger FVC, morningness, sunburn risk). 🧑🦲🫁🌞
For other traits, we observed no/little directional bias.
7/
For some traits, Neanderthal variants contribute a directional effect (balding risk, younger menopause, larger FVC, morningness, sunburn risk). 🧑🦲🫁🌞
For other traits, we observed no/little directional bias.
7/
To contextualize these results, we propose a model that links these above results:
1. observed patterns of heritability and
2. direction of effect
to hypotheses about the history of selective pressures on introgressed haplotypes.
8/
1. observed patterns of heritability and
2. direction of effect
to hypotheses about the history of selective pressures on introgressed haplotypes.
8/

I think I best describe the intuition behind these scenarios with examples in this post: natureecoevocommunity.nature.com/posts/uncoveri…
For example, consider the differences in potential pressures on introgressed variants associated with sunburn(skin color) vs. diabetes(glucose homeostasis)
9/
For example, consider the differences in potential pressures on introgressed variants associated with sunburn(skin color) vs. diabetes(glucose homeostasis)
9/

The relationship between complex trait heritability and evolution/selection is so complicated and interesting! I spent a lot of time trying to envision different scenarios. So for fun/learning, I built an interactive app: neanderthal-heritability.herokuapp.com with @plotlygraphs @heroku
10/
10/
These models are an oversimplification of a lot of complex environmental/genetic/demographic factors. However, we hope our discussion/model provides useful context for our results and a framework for future work. Feedback and thoughts welcome!
11/
11/
Notably, our work only quantifies contribution of common variants in European populations. We anticipate more genetic data from non-European populations, new statistical methods, and future simulations will continue to provide a window of understanding into our past! 🌏👩💻
12/
12/
This work would not have been possible without an amazing community and some really thoughtful reviewers (check out new results since our @biorxivpreprint !) Shout out to:
@capra_lab
@DaRinker
@VUMCgenetics
@ACCREVandy
@VanderbiltMSTP
@UCSF_BCHSI
@UCSF_Epibiostat
13/13
@capra_lab
@DaRinker
@VUMCgenetics
@ACCREVandy
@VanderbiltMSTP
@UCSF_BCHSI
@UCSF_Epibiostat
13/13
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh