I'm super proud to share #Lithuania's #SARSCoV2 genomic surveillance programme's 1st paper, published today in @NatureComms. It's about a worrisome lineage called B.1.620 discovered in 🇱🇹, across Europe & traced to central Africa. Here's a 🧵about it nature.com/articles/s4146… 1/15 

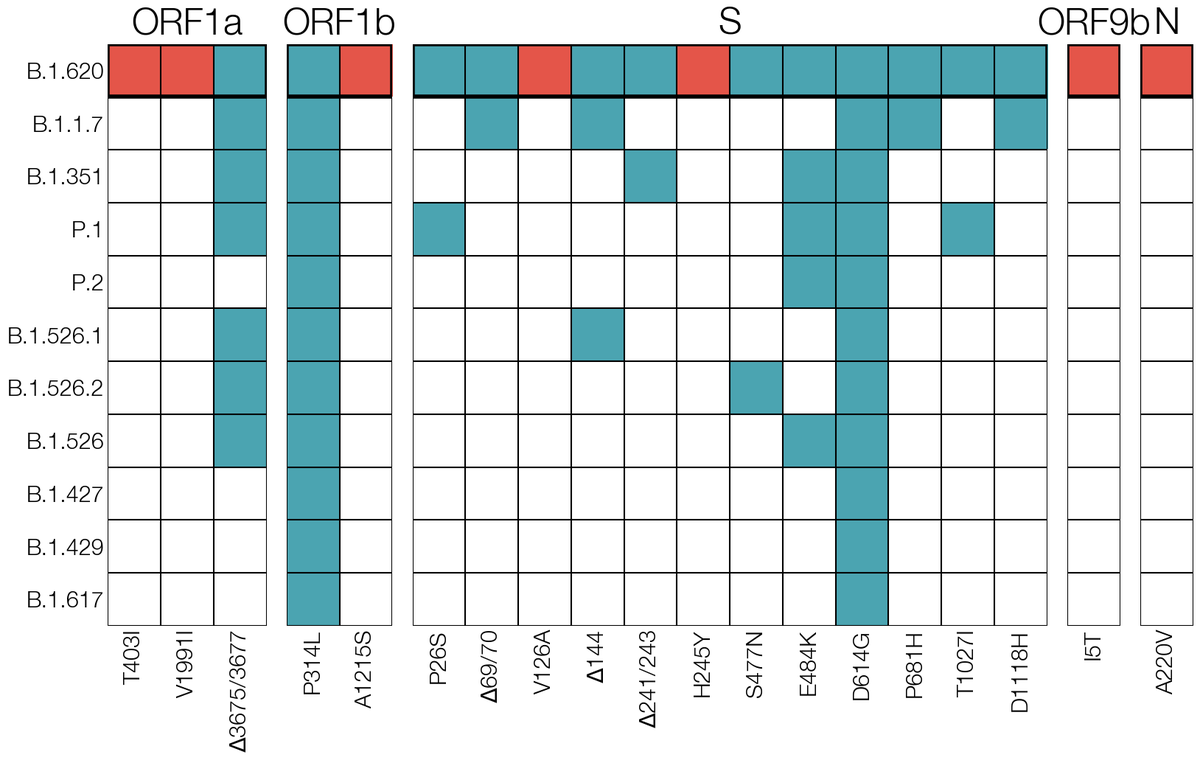
In April 2021 when Delta wasn't a "thing" yet & B.1.1.7/Alpha was in decline thanks to vaccines we suddenly discovered a diverged #SARSCoV2 lineage with many VOC-like mutations which had no sampled relatives & which was suddenly appearing across Europe. We were very alarmed. 2/15 

On week of April 5 @Ingridole noticed 2 #SARSCoV2 genomes flagged by nextclade for having very long branches. They were related & the long branch encompassed a cocktail of many notable mutations VOCs of the day (Alpha, Beta, Gamma) carried. It looked like a parody of a VOC. 3/15 
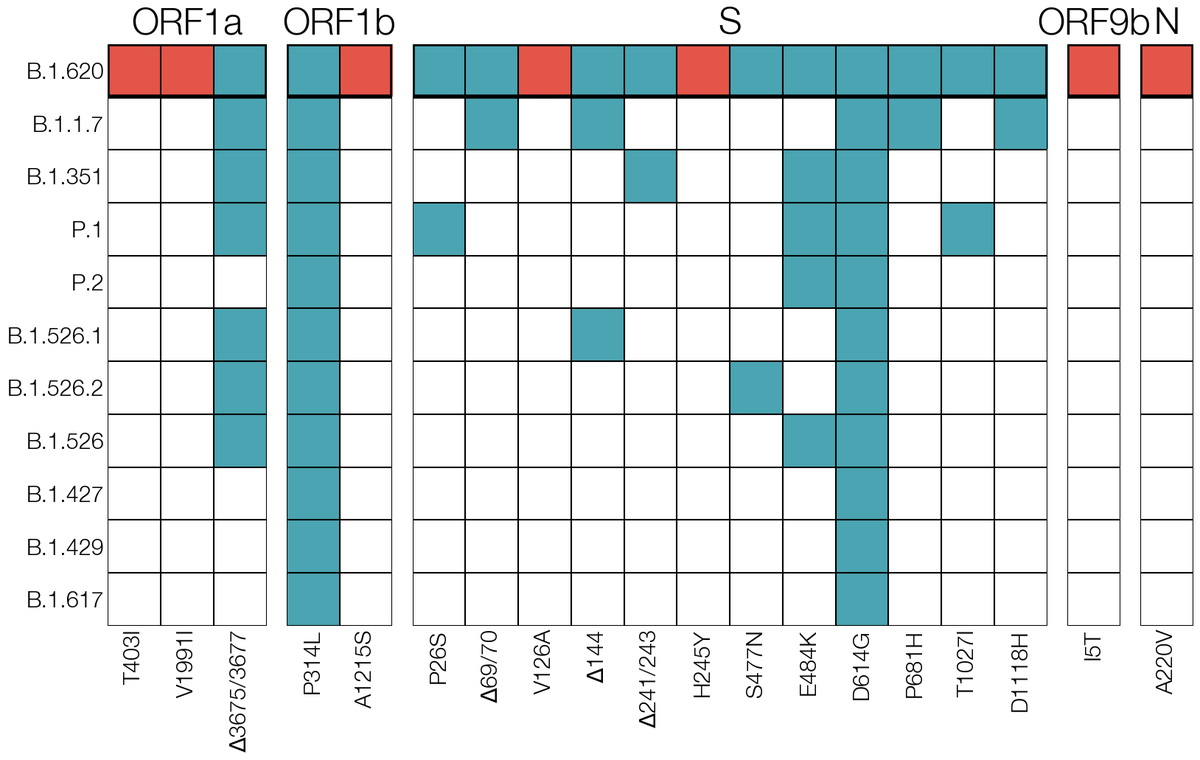
We thought contamination first, but Beta & Gamma lineages weren't circulating in-country & we had two samples, not a singleton. Recombination wasn't it either since regions between its Alpha-like mutations were missing mutations Alpha had. This unfortunately looked legit. 4/15 
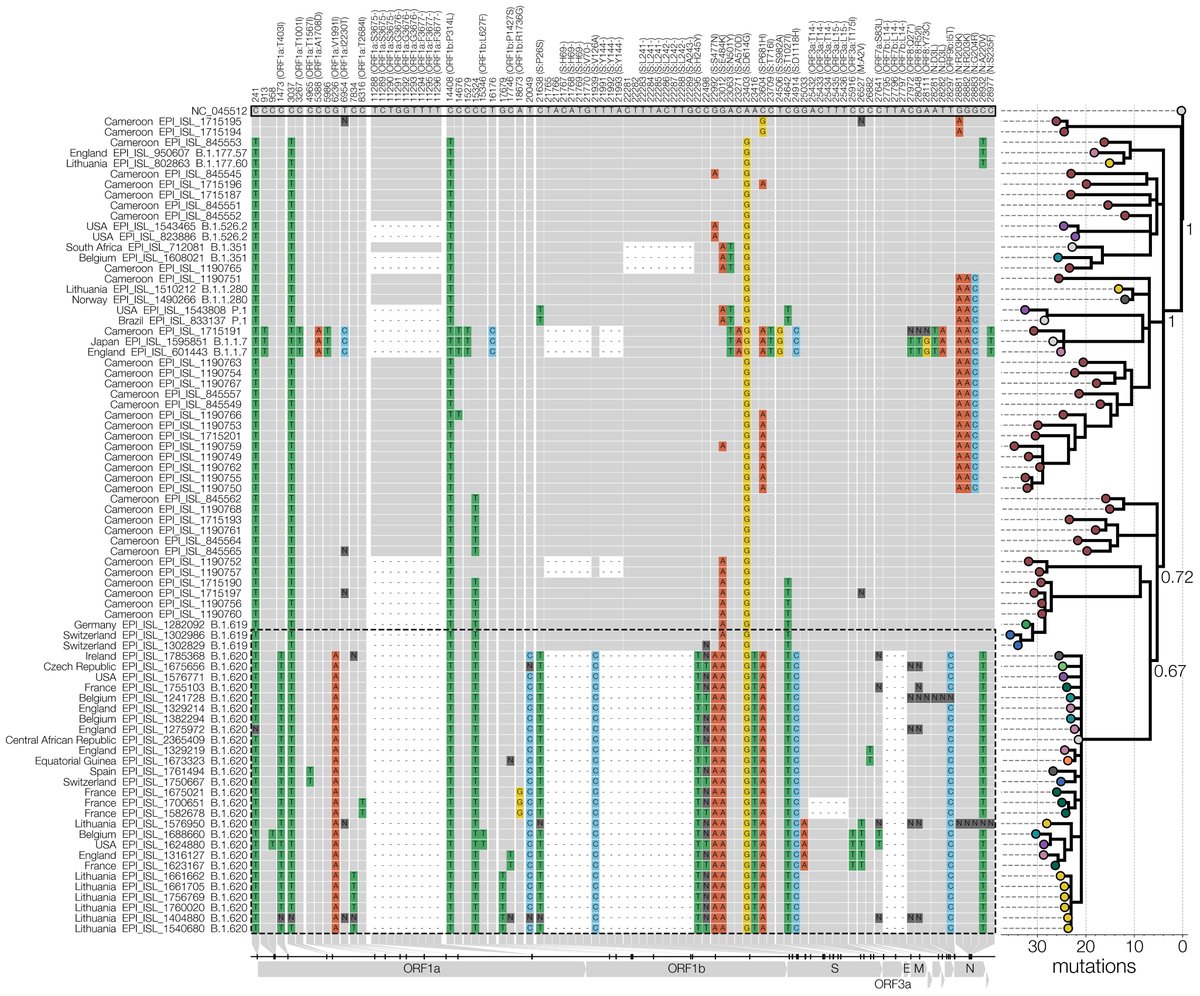
Worse yet, we discovered that another team in Lithuania saw this lineage a week earlier but missed it because most of its genomes were being misclassified by pangolin as B.1.177.57, an innocuous lineage if you ignore E484K & don't know what mutations B.1.177 has by heart. 5/15
Searching on GISAID for some of its mutations we discovered more B.1.620 genomes dispersed throughout Europe. All in small numbers, mostly unrelated to each other, even if found in the same country. It wasn't common & didn't look endemic so we looked for potential sources. 6/15 
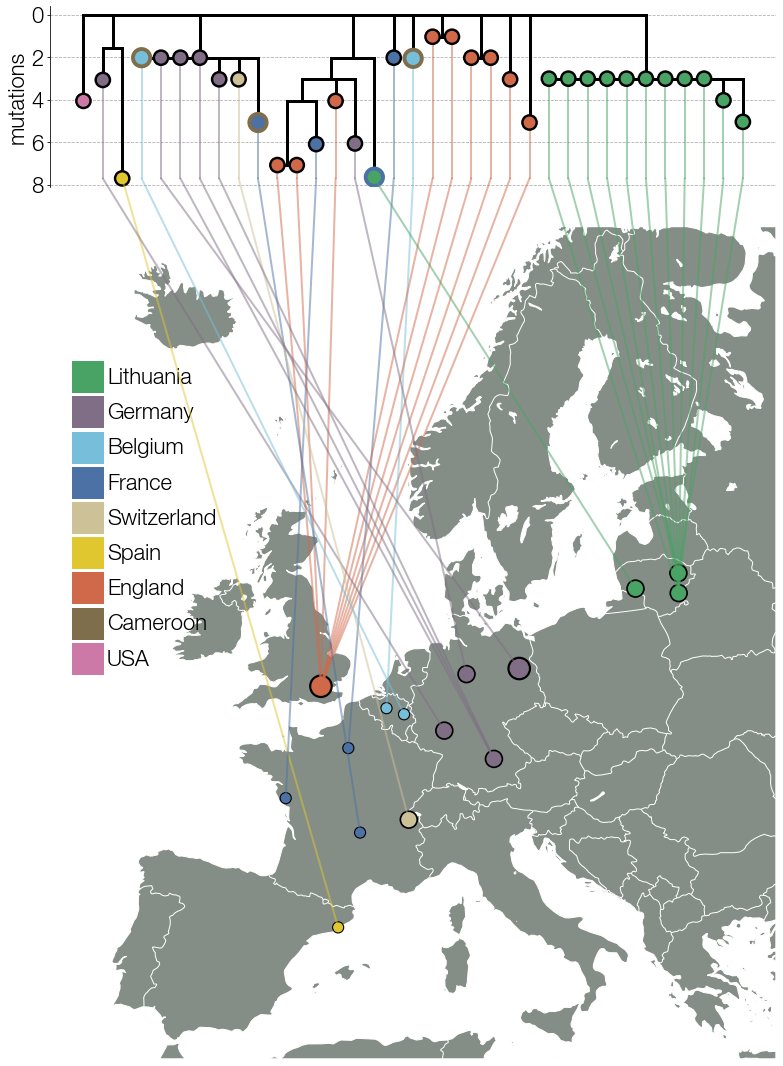
Our suspicions were strengthened when we noticed that the earliest (then known) case was a traveler coming from Cameroon. Contacting submitters of B.1.620 genomes to GISAID found even more Cameroon travelers. To top it off we unexpectedly received some smoking-gun data. 7/15 
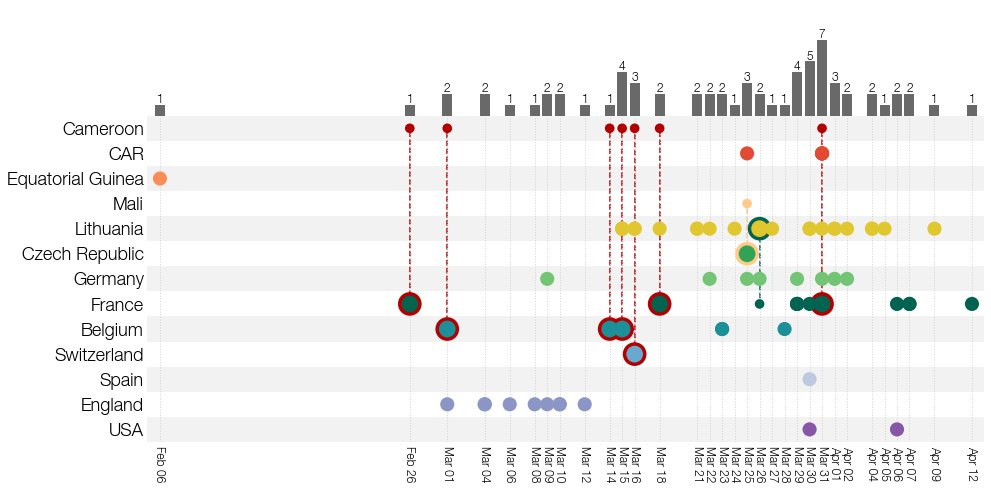
Thanks to @GuyBaele & Séb Calvignac-Spencer we got genomes of this lineage from Central African Republic near the border with Cameroon. These were closer to the basal genotype B.1.620 would have had. Everything fit & we finally had a convincing story we just had to write up. 8/15 
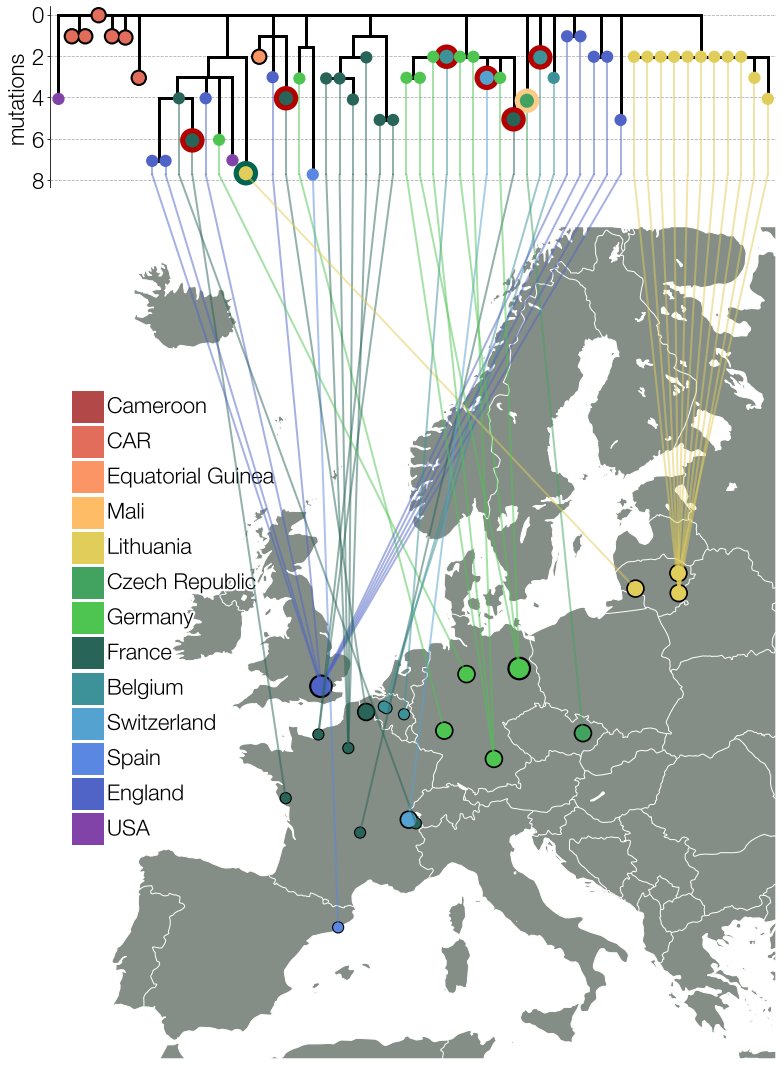
It took us 23 days from 1st observation to submitted manuscript. During this time our lineage X got designated B.1.620, South Korea reported a big outbreak of it & many genomes from CAR showed up on GISAID. But as the B.1.620 story was getting interesting Delta happened. 9/15 
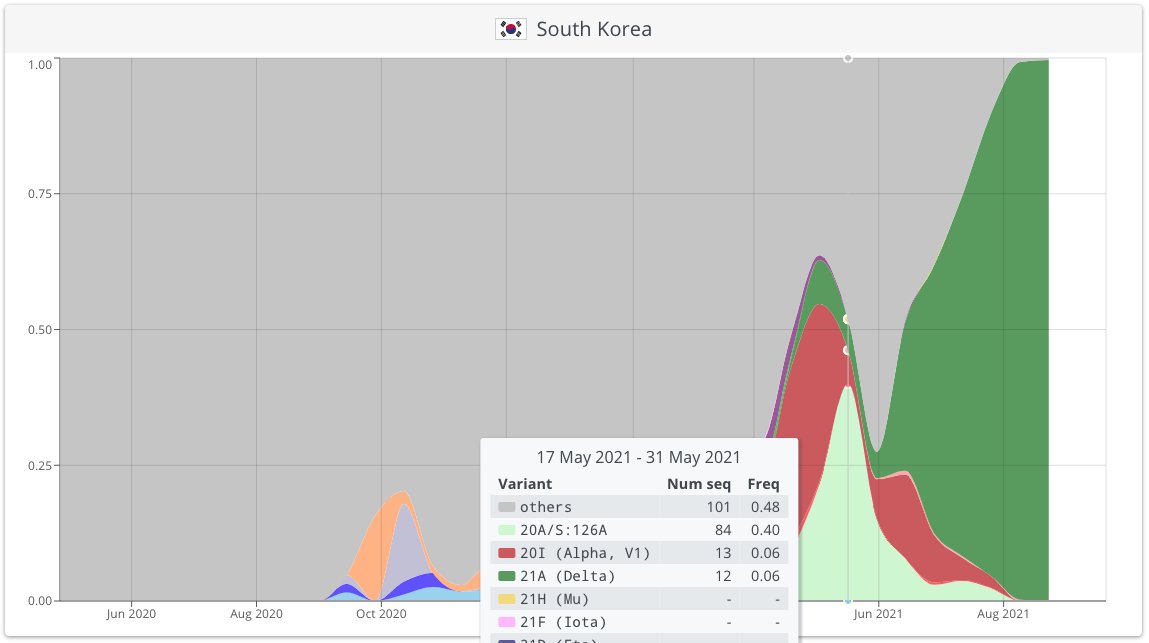
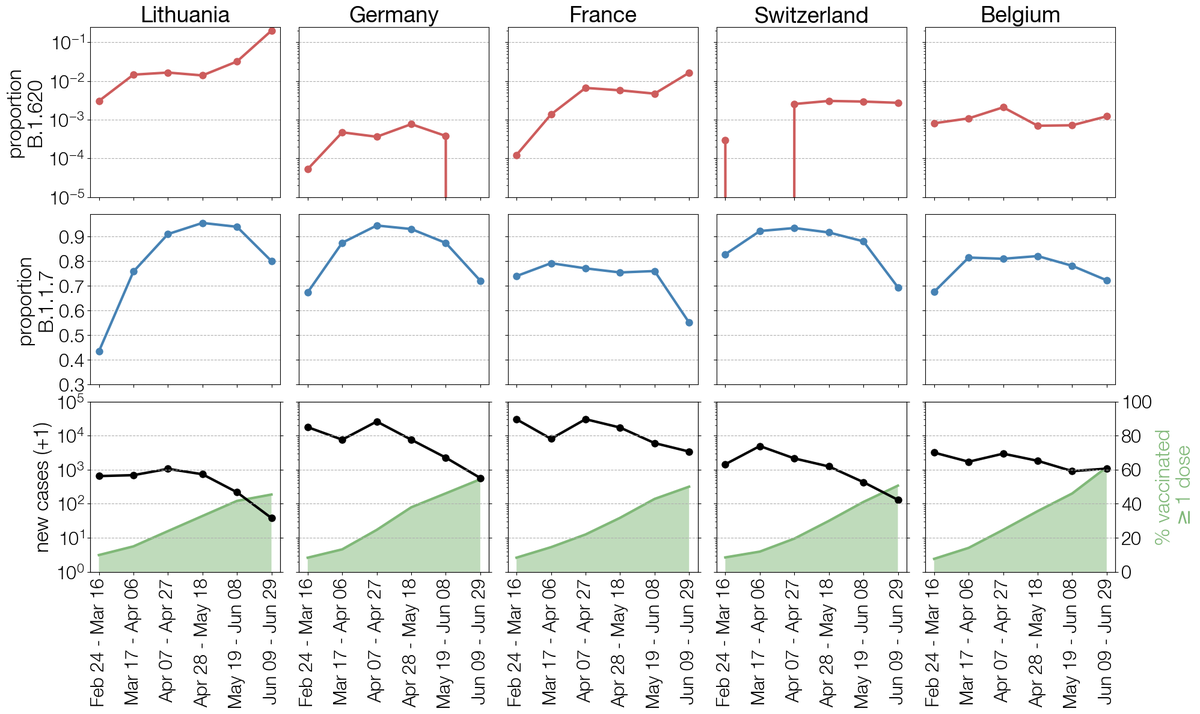
During peer-review in June we saw signs that B.1.620 had an advantage over Alpha (probably via antigenic differences) but when Delta took over Europe in July/August somehow B.1.620 just shriveled up. B.1.620 was last seen in Lithuania Jun 24 & ever in Switzerland Sep 06. 10/15 
Though likely extinct now, B.1.620 was a lesson. Failure made flesh. Faltering response (in central Africa & Lithuania) can be excused but vaccine hoarding & no genomic surveillance assistance in Africa backfired predictably. Viruses get to be global but compassion doesn't. 11/15
In articles about this study's preprint @meredithwadman, @zulfikarabbany, & @SmritiMallapaty laid these failures bare. Even now we in Europe get to plan what we'll do after "all of this" is over (depending on vaccination level) while some still bury their dead en masse. 12/15 



Ultimately the best thing to come out of the pandemic for me was meeting some truly inspiring people in Lithuania: @Ingridole, @MGabrielaite, Jonas Bačelis, Daniel Naumovas, Dovilė Juozapaitė, Rimvydas Norvilas, Aistis Šimaitis, & Emilija Vasiliūnaitė, to name just a few. 13/15
I also got to work with some old friends (@GuyBaele, @barneypotter24, @joostsnijder), met new ones (@SamLHong, @PlacideMbala, Kamran Khan, Séb C-S), & saw African scientists doing a great job locally. It wasn't easy work but I'm glad I had a great team to collaborate with. 14/15
To conclude, even though B.1.620 is probably dead the circumstances that let it happen are still alive. C Africa seems far away from Europe but the world we live in simply doesn't work that way anymore. My consolation is that I got to work with great people because of it. 15/15
• • •
Missing some Tweet in this thread? You can try to
force a refresh