Just out - the rise and fall of SARS-CoV-2 lineages in England.
In the last 1.5yrs the UK has been a bell weather for SARS-CoV-2 evolution and genomic epidemiology thanks to the data sequenced by @CovidGenomicsUK and @sangerinstitute.
Let's recap. >>
nature.com/articles/s4158…
In the last 1.5yrs the UK has been a bell weather for SARS-CoV-2 evolution and genomic epidemiology thanks to the data sequenced by @CovidGenomicsUK and @sangerinstitute.
Let's recap. >>
nature.com/articles/s4158…
As any virus, SARS-CoV-2 accumulates mutations and undergoes an evolutionary journey where fitter variants succeed. Most mutations are neutral and enable us to define lineages, which derive from a single ancestor and share all its mutations. By now there are >1000 lineages. >>
As new variants emerge all the time it is important to characterise their behaviour as soon as possible and an essential question is whether one variant has a growth advantage over others, as this may change the future course of the epidemic. >>
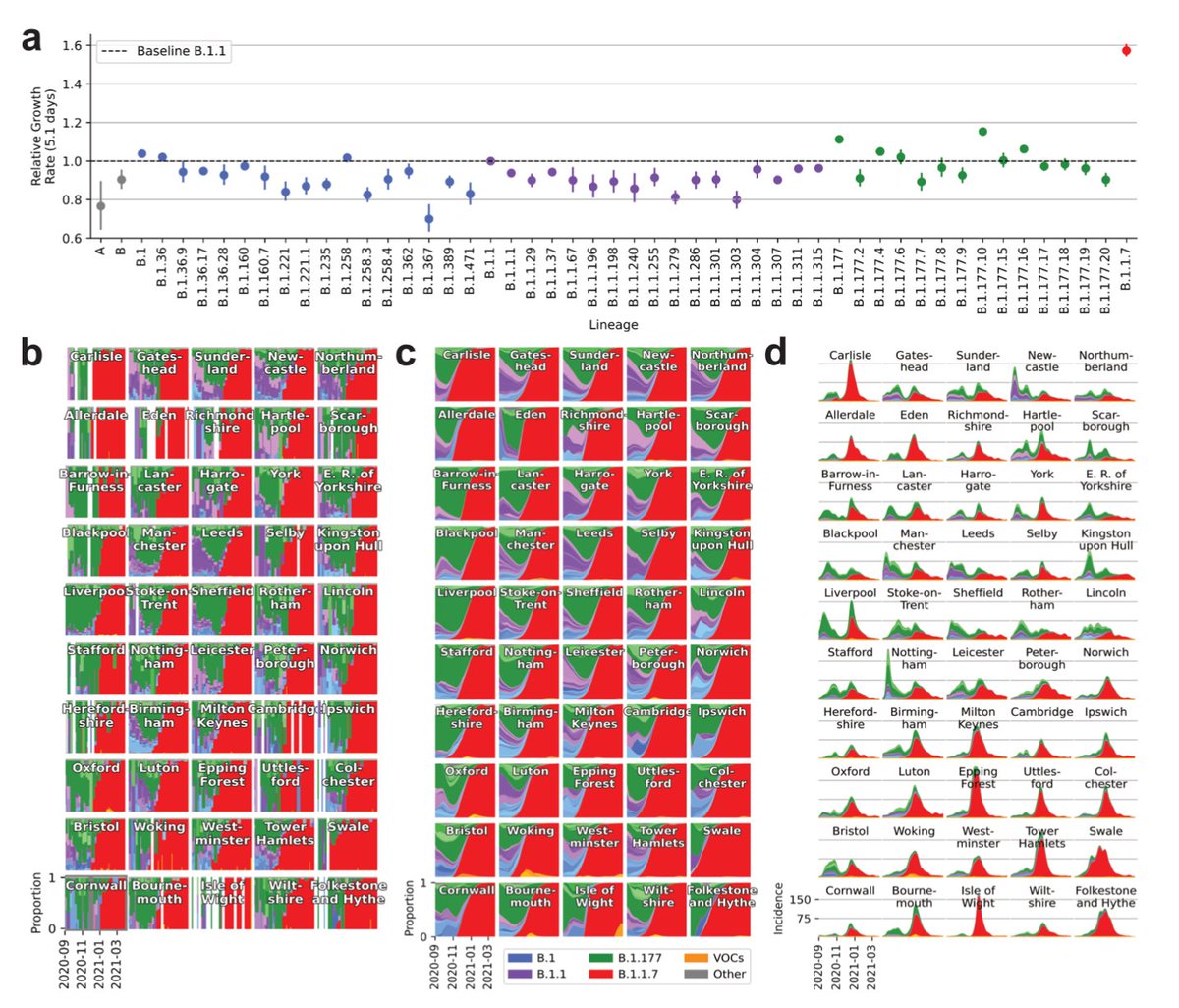
During the first wave in the spring ‘20 @erikmvolz observed that a lineage with a spike mutation D614G grew spread around 15% faster than other lineages. This lineage was termed B. >> sciencedirect.com/science/articl…
Over the summer ‘20 another lineage termed B.1.177 floated across Europe, largely facilitated by holiday travel as described by @firefoxx66 . From the ‘177’ you can already guess that a great number of lineages had been defined by then. >> nature.com/articles/s4158…
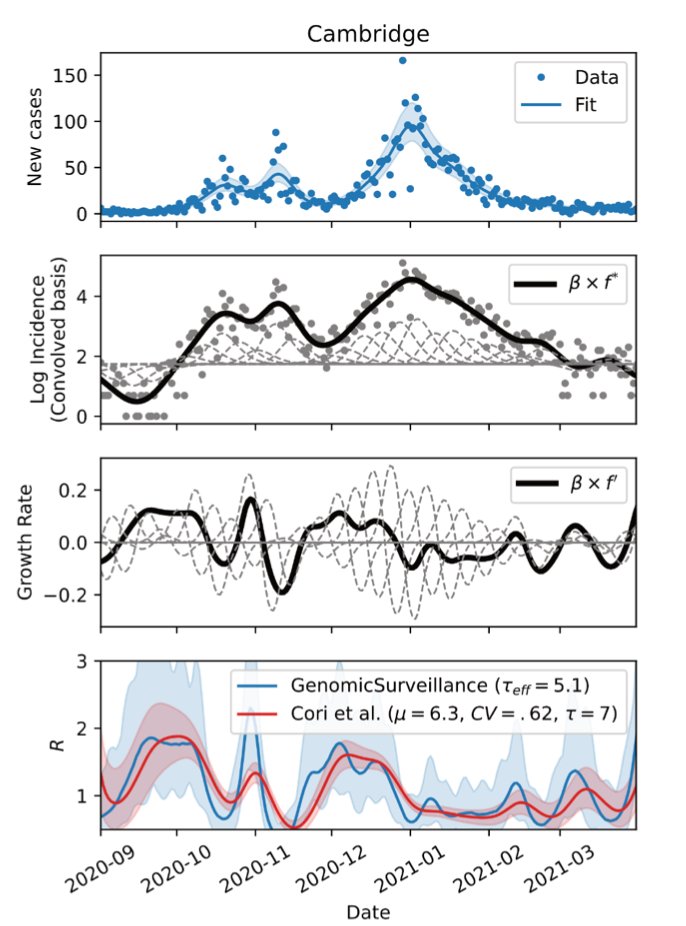
This is also where our analysis begins, because by the time the genomic surveillance sequencing @sangerinstitute was fully up and running. Essentially @jcbarret and colleagues sequenced ~10% of all positive specimens, collected from testing the wider population in England. >> 

In order to keep track of the variety of existing lineages spreading in different places @harald_voeh in my group build a model capable of tracking 71 variants across 315 English local authorities and calculating their growth rate and incidence. >> github.com/gerstung-lab/g… 

During the fall ‘20 B.1.177 continued to rise slowly, consistent with a 10% growth advantage. That was nothing that people were too worried about. >> 



At the same time, however, a new variant emerged in the U.K., termed B.1.1.7 (later termed Alpha by the WHO). It started in the South East of England and spread across the entire nation. It took some time to piece this information together. Were people there not following rules? 

At this stage watching how Alpha spread in a side by side comparison in each of 315 local authorities showed that it possessed a consistent growth advantage of around 50%. What’s more is that this enabled it to grow during a lockdown when other lineages _declined_. >> 

Unfortunately, the full extent of the Alpha threat wasn’t apparent when England opened up in December (cases had been falling as Alpha numbers were still low) and the nation experienced a catastrophic surge leading the NHS close to collapse. >>
The insight that Alpha’s growth advantage was sufficient to beat the November lockdown led to a more extensive one in January ‘21. These measures, paired with immunity from infection and increasingly vaccination, curbed Alpha’s spread. >> 

As Alpha had a ~50% growth advantage, falling Alpha numbers translated to an even more rapid decline of B.1.177 and other variants present in the summer of ‘20. In fact, most of these lineages were _eliminated_ in the spring of ‘21. >> 

Yet another threat emerged. Across the globe a number of lineages surfaced which possessed the ability to dodge recognition by neutralising antibodies, including the B.1.351/Beta and P.1/Gamma variants of concern. A common feature was the immunity evading E484K mutation. >>
E484K variants indeed entered the country in considerable numbers, but couldn’t quite hold their ground due to existing restrictions and potentially targeted measures. Still, collectively they slowly rose to ~1.5% of cases in April ‘20. >> 



Then something surprising occurred once more. A large wave had gripped India and was associated with a new variant, B.1.617. But the data was unclear. Genomic data was patchy, B.1.617 appeared to have existed for a long time and it didn’t contain any of the usual mutations. >>
Unfortunately B.1.617 and especially its sublineage B.1.617.2/Delta had already got a foothold in the UK, due to extensive links between the countries, and numbers began to climb rapidly. In early May it became clear that Delta had a serious growth advantage over Alpha, ... >> 



... but how much exactly remained difficult to assess because of the rate of introductions. When the dust settled it was clear that Delta possessed at least 60% growth advantage over Alpha, which created a large surge of cases, even though Alpha cases continued falling. >> 

The mechanisms of Delta’s speed-of-light-like spread are not fully understood yet, and are likely a consequence of its effortless cell entry and ability to dodge prior immunity. >>
Delta’s huge transmissibility has profound consequences. Even though vaccines remain highly effective against infection and esp. severe disease, the elevated herd immunity threshold 1-1/R means that vaccination alone won’t eliminate Delta. >>
https://twitter.com/adamjkucharski/status/1415587167863283713
Instead there will be a transition into an endemic state where circulating variants periodically infect individuals whose immunity has waned or where immunity doesn't protect as well against a particular new variant. >>
While this may sound shocking, this is the situation with many other respiratory viruses. Prior immunity will greatly reduce the severity of these subsequent infections. >>
https://twitter.com/BallouxFrancois/status/1411697682897326081?s=20
But there are also a number of unknowns for SARS-CoV-2, such as how quickly immunity wanes (with and without boosters), the rate of antigenic drift facilitating immune escape etc. There could also be further adaptative evol. increasing transmissibility. >>
https://twitter.com/trvrb/status/1448297954306150401?s=20
Where are we at the moment?
So far, no serious contender to Delta has emerged in our data.
There are a great number of Delta sublineages (AY.x), which exhibit moderately different growth patterns and are extensively circulating in England and globally. >>
So far, no serious contender to Delta has emerged in our data.
There are a great number of Delta sublineages (AY.x), which exhibit moderately different growth patterns and are extensively circulating in England and globally. >>
In the mean time, after only a year, Alpha seems to be history in England.
While the exact future is impossible to predict, a pattern of rising and falling lineages seems likely. >>
While the exact future is impossible to predict, a pattern of rising and falling lineages seems likely. >>
https://twitter.com/MoritzGerstung/status/1443218609665449989?s=20
This work was a fantastic collaboration with many of the brightest minds. Thanks also to all the other hard working scientists who have sacrificed so much for the world to stay on top of the rapidly evolving epidemic and whose work has shaped ours. @jcbarret @meera_chand 

And perhaps another thing to add is that genomic surveillance data used in our study and updated every week since continues to be available at covid19.sanger.ac.uk, developed by @theosanderson, Richard Goater and others.
• • •
Missing some Tweet in this thread? You can try to
force a refresh