Picking up where @rita_rebollo left, we spent a couple of years annotating and analyzing the rest of the rice #weevil #genome (ok, the 15% left after removing repeats)! All info presented here is on the huge Additional file 2 in @BMCBiology doi.org/10.1186/s12915… 1/n
@VargasChavezC and @gabaldonlab performed a comprehensive #phylogenomic analysis of the rice #weevil and found that this insect has a high gene expansion rate when compared to other beetles. 2/n 

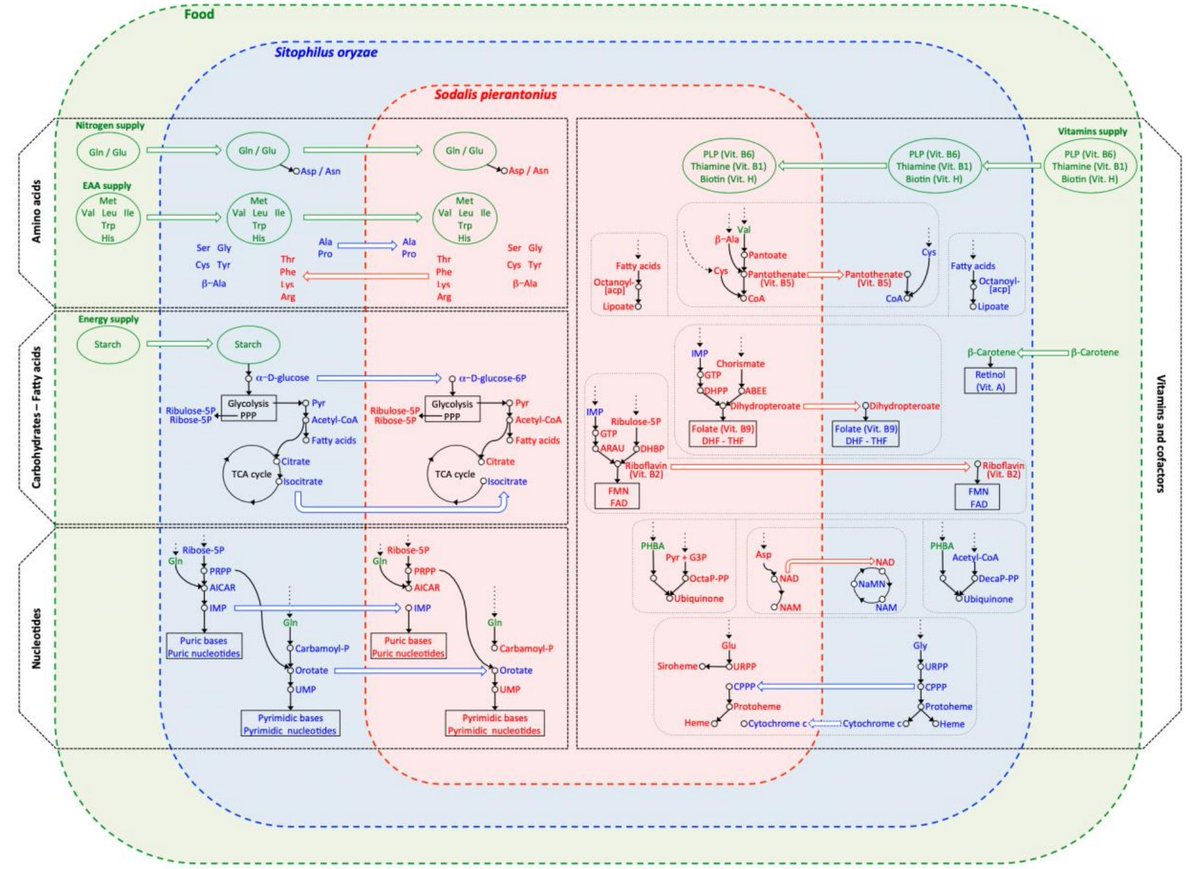
With the invaluable help of the SymT group from @INRAE_UMR_BF2I we have also reconstructed the #metabolic pathways from both #symbiotic partners. Specific losses of enzymes in the #weevil genome are limited while its #endosymbiont has suffered much more degradation. 3/n
@VargasChavezC and I annotated lots of gene families including digestive enzymes, cuticle proteins, detoxification and #insecticide resistance related genes. 5/n
And thanks to the help of our collaborators we had the opportunity to annotate developmental genes (@CallaertsLab from @KU_Leuven) and odorant receptors (@EmmaJoly8, N. Montagné et @CamilleBurgh from @iEESParis) 6/n
The analysis of #immunity related genes was done at @INRAE_UMR_BF2I. @cmonegat and @AnnaZaidmanRemy coordinated the work of our former #PhD students @VargasChavezC @Justin_Maire @flmasson and A. Vigneron. 7/n
They found that despite the interaction with S. pierantonius, the rice #weevil genome encodes the canonical genes involved in the 3 main #antimicrobial pathways Toll, Imd, and JAK-STAT. 8/n 
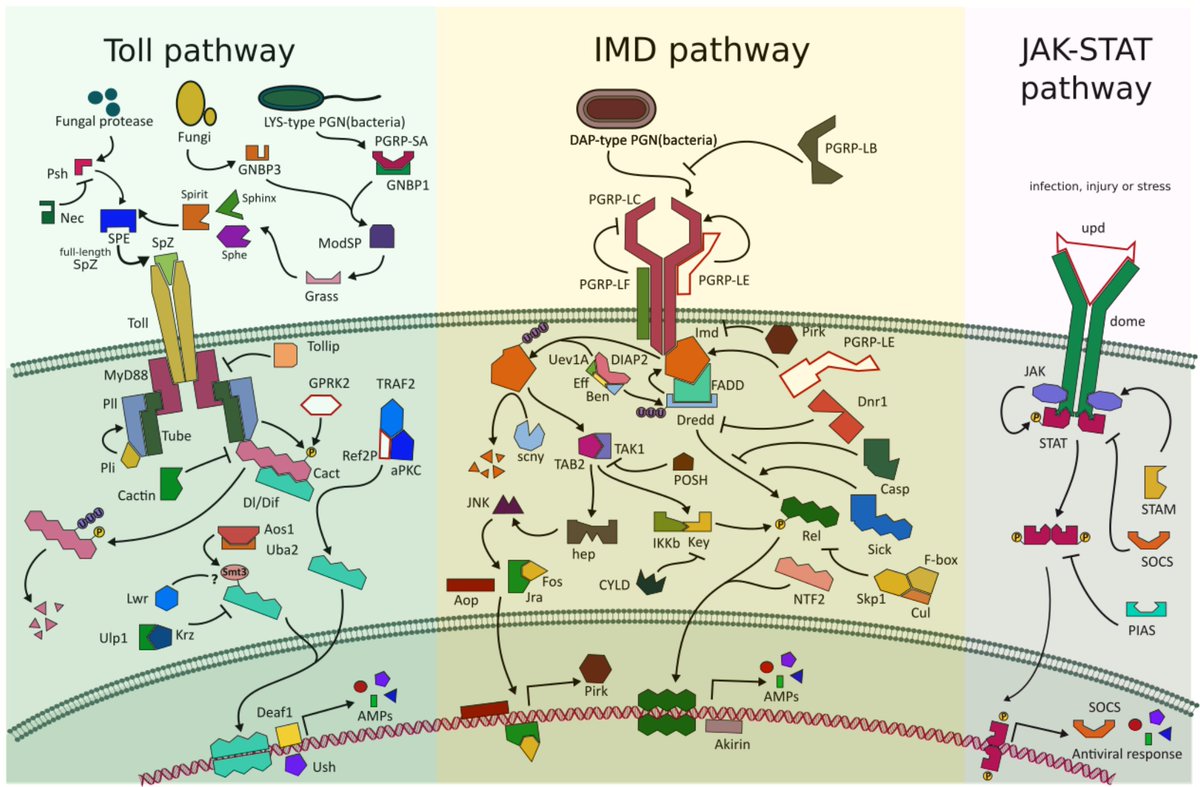
The Imd pathway is not only present but it is also functional and has evolved molecular features necessary for endosymbiont control and host immune homeostasis. As seen in doi.org/10.1186/s40168… and doi.org/10.1073/pnas.1… 9/n
On #epigenetics @rita_rebollo, A. Vallier, @GAUTIERRichard, @VargasChavezC & co found only DNMT1 & 2 (not 3) and around 1% of #CpG methylation. They also annotated the #piRNA genes, and albeit the presence of SIWI, most piRNA-related genes are overexpressed in germ vs soma 10/n
I would like to end this thread by thanking A. Heddi and @amparo_latorre for integrating me in the middle of their journey and for their strong support. 11/n
A big thanks also to @VargasChavezC for his incredible contribution, so challenging given the complexity of the genome. 12/n
Finally, I thank @rita_rebollo @clementgoubert and @TreepVieira for the time spent to analyze this genome full of repeats. You, guys, can be proud of what you have done here! 13/13
• • •
Missing some Tweet in this thread? You can try to
force a refresh