An update on 🦠 SARS-CoV-2 variants in England.
Today: AY.4.2.1, AY.98 and other introductions.
TLDR: It seems to be ok.
Today: AY.4.2.1, AY.98 and other introductions.
TLDR: It seems to be ok.

Background: The list of Delta sublineages has gotten longer – more than 121 AY.x lineages + sublineages.
This is thanks to the virus hunters @PangoNetwork spotting epidemiologically relevant variants across the 🌍.
These appear as colored branches at cov2tree.org
This is thanks to the virus hunters @PangoNetwork spotting epidemiologically relevant variants across the 🌍.
These appear as colored branches at cov2tree.org

In England, @CovidGenomicsUK operates a very systematic sequencing programme, which allows to detect & characterise the spread of new lineages early.
Every Monday @jcbarret and his team release data from around 25,000 samples (~10% of cases) on covid19.sanger.ac.uk
Every Monday @jcbarret and his team release data from around 25,000 samples (~10% of cases) on covid19.sanger.ac.uk
In order to shorten the 2-3 weeks between lineage spotting and annotation, @theosanderson has developed a tool that rapidly assigns genomes to the very latest lineages.
(I mention this because the data is slightly ahead of Monday's release using official annotation.)
(I mention this because the data is slightly ahead of Monday's release using official annotation.)
This latest, provisional data includes a number of new lineages down to AY.120.2.1.
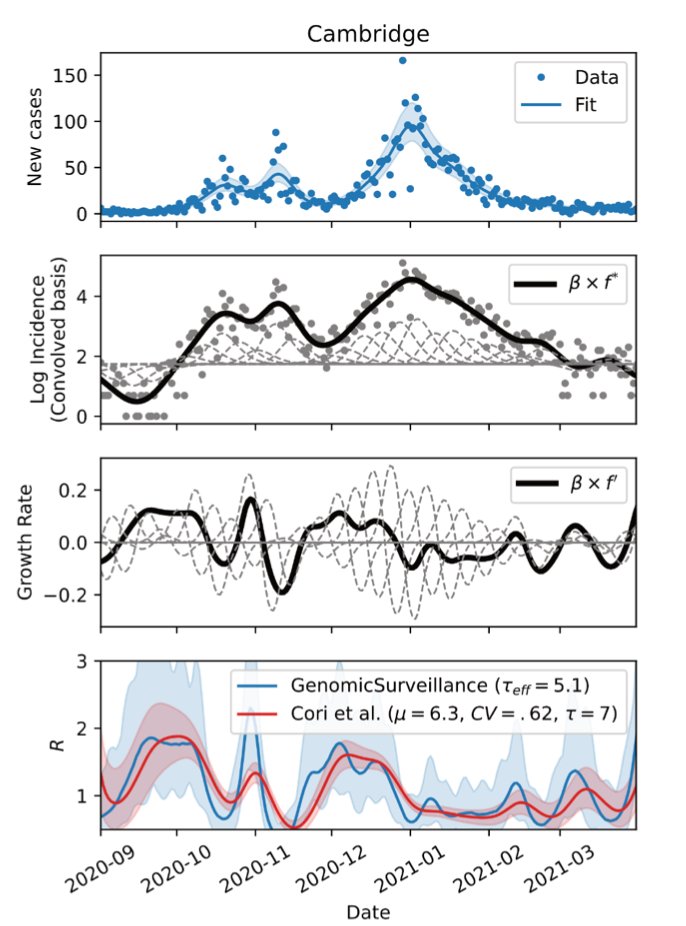
We have been using the model by @harald_voeh to track the fate of 278 lineages. nature.com/articles/s4158…
We have been using the model by @harald_voeh to track the fate of 278 lineages. nature.com/articles/s4158…

The first lineage to mention is AY.4.2.1, the purple emerging clade within the green AY.4.2 branch, spotted by @CorneliusRoemer.
It comprises ~3400 samples by now and is found in all English regions, comprising between 1% and 4% of samples in the week ending 6 Nov.
It comprises ~3400 samples by now and is found in all English regions, comprising between 1% and 4% of samples in the week ending 6 Nov.

AY.4.2.1's growth was, on average, ~1% faster per day compared to AY.4.2. In some regions it was about the same as AY.4.2.
👩🔬: AY.4.2.1 has spike mutation V36F, which could be a hint of a mechanism. But r=1% is within the typical range of fluctuations, so it's inconclusive.


👩🔬: AY.4.2.1 has spike mutation V36F, which could be a hint of a mechanism. But r=1% is within the typical range of fluctuations, so it's inconclusive.



Another new lineage is AY.98 (green), which is rather numerous (n=11k samples) now comprising between 2%-5% of samples.
👨💻: AY.98 is in fact a rather old branch of Delta and has been added mostly to complete the taxonomy.
👨💻: AY.98 is in fact a rather old branch of Delta and has been added mostly to complete the taxonomy.

It has been increasing slowly in all regions except the North East, where it's stagnant at higher levels. 🤔
🧑🏫: AY.98 was among the first founders of the Delta wave in N/E, reaching high numbers by chance. It now slowly diffuses to other regions until the frequencies are equal.


🧑🏫: AY.98 was among the first founders of the Delta wave in N/E, reaching high numbers by chance. It now slowly diffuses to other regions until the frequencies are equal.



Lastly, there are also many currently uncharacterised variants across the 🌍.
As the pandemic accelerates in regions such as Eastern Europe, variants from such places also appear in the UK in increasing numbers.
As the pandemic accelerates in regions such as Eastern Europe, variants from such places also appear in the UK in increasing numbers.

One example is the pink branch below, defined by Orf3a:202L. It doesn't have a pango lineage name yet.
The branch is prevalent in Eastern Europe (🇩🇪: 6-10%, 🇵🇱: ~10%, 🇹🇷 20-30%). But the relative proportions there are fairly stable, arguing against a large growth advantage.
The branch is prevalent in Eastern Europe (🇩🇪: 6-10%, 🇵🇱: ~10%, 🇹🇷 20-30%). But the relative proportions there are fairly stable, arguing against a large growth advantage.

In England Orf3a:202L is mostly found in London, where it rose noticeably to around 80 cases (2%) per day. These numbers are comparable to AY.4.2.1; the growth was even faster. 😳
👩💻: The increase mostly reflects the rising cases elsewhere rather than an inherent advantage.
👩💻: The increase mostly reflects the rising cases elsewhere rather than an inherent advantage.

So overall, the situation in England seems to be in a fluent state, as expected.
The spread of most lineages reflects the stochastic and complex domestic and international dynamics of the pandemic.
AY.4.2.1 may have a very small growth advantage, but I wouldn't bet on it yet.
The spread of most lineages reflects the stochastic and complex domestic and international dynamics of the pandemic.
AY.4.2.1 may have a very small growth advantage, but I wouldn't bet on it yet.
• • •
Missing some Tweet in this thread? You can try to
force a refresh