
Time for the latest @nanopore technology updates. Up next in the auditorium: an Update from the Oxford Nanopore team. #nanoporeconf 

Please note, we invite you to read the disclaimer in the slides
CB: More information on Clive’s previous technology updates:
Next up is James Clark (JC). For a platform update. #nanoporeconf
JC: Our technology has been built on firm foundations and has matured significantly over the past couple of years. #nanoporeconf
JC: we are bringing together years of innovation to showcase one sensing platform for all biological analyses #nanoporeconf
JC: Algorithm & chemistry improvements keep pushing up accuracy. Kit 14 alongside the R10.4.1 #nanopore has been shown to achieve 99.6% modal raw read accuracy. Shown to achieve 99.2%, whilst maintaining speeds of 420 bases per second. #NanoporeConf
JC: We plan to move all nanopore platforms over to the R10 series of nanopores. #nanoporeconf
JC: Users can tune their #PromethION run to optimise their output. R10.4.1 outputs looking nice, but still tuning up the new pore. #nanoporeconf 

JC: The Mediator is the limiter. More Mediator in the well gives consistent currents for longer resulting in higher output per channel during sequencing and long-life flow cells. #nanoporeconf
JC: Our first #PromethION 2 (P2) was shipped last week. See here:
https://twitter.com/DrT1973/status/1525150138506702849#nanoporeconf
JC: Register your interest for P2 here: nanoporetech.com/products/p2 #NanoporeConf
JC: we still have some unfinished business such as improving our flow cell design, lowering sample volumes, and improving robustness to ensure ease of use. #nanoporeconf
JC: Manufacturing is scaling up volumes as demand increases. We have dedicated teams to ensure quality of inputs and processes. This is a continuous process which leads to higher yields, less waste, more robust flow cells. #nanoporeconf
JC: new ASICs are being assessed in-house. The next generation of ASIC is designed for low noise & low power to enable the development of new product formats. #nanoporeconf 

JC: demonstrating the new ASIC #nanoporeconf 

Next up, Lakmal Jayasinghe (LJ) with an update on chemistry. #nanoporeconf
LJ: we can drive *output* using numbers of nanopores; driven by ASIC design, chemistry, membrane, nanopores; also speed of sequencing (faster for higher output), and a well prepared, well-loaded library to maximise ‘on’ time. #nanoporeconf
LJ: A quick update on modal single-molecule accuracy. Older baselines (Kit 9/10) have lower accuracies but fast motor (420 bases/sec). The Kit 12 has Q20+ accuracies but slow motor (250 bases/sec) – there was a need to break the output/accuracy trade-off. #nanoporeconf 

LJ: The new chemistry (Kit 14) alongside a new nanopore (R10.4.1) has been shown to achieve 99.2% modal raw read accuracy, whilst maintaining speeds of 420 bases per second; for similar data yields to the current high-yield R9.4.1 flow cells. #nanoporeconf 

LJ: Kit 14 will allow users to tune their accuracy & output needs by modifying run conditions — programmable in the software. Early work has shown 99.6% raw read accuracy can be achieved when run at 28 degrees. At 34 degrees 99.2% is possible whilst generating a much higher yield 

LJ: The duplex complement strands are paired informatically. Duplex is also improved with the new chemistry and users are now able to achieve ~Q30 modal accuracy following kit updates. #nanoporeconf 

LJ: R10.4.1 has improved homopolymer accuracy compared to R9.4.1. The next generation will have even longer reader heads to further improve homopolymer accuracy. #nanoporeconf 

Next up, a data update from Stuart Reid (SR) #nanoporeconf
SR: Reminder there are several ways to improve the accuracy of @nanopore devices. We have driven performance improvement through continuous innovation in our chemistry, platform & training of basecalling models #nanoporeconf
SR: Accuracy means different things for different applications – which is most important to you? #nanoporeconf
SR: Raw read ‘simplex’ accuracy with Kit 14 and R10.4.1 nanopore modal accuracy 99.2 % (Q21) at 400 basepairs/sec. #nanoporeconf 

SR: We will be introducing a choice of run conditions: “Accuracy” at 260 bps for 99.6% raw read accuracy, “Default” and “Output” at 520 basepairs/sec increasing the output of a single #PromethION Flow Cell to 307 Gb. #nanoporeconf
SR: results from ultra-long duplex reads and HG002 dataset with 260 bps condition. Modal accuracy ~Q30. Longest, Q30 duplex read: 260 kbases. Longest Q40 duplex read: 144 kbases. Longest read that perfectly aligns to a human reference: 72 kbases #NanoporeConf 

SR: You get complete, contiguous, accurate genomes with #nanopore data - Q50+ bacterial assemblies with modest 10-20X coverage.
SR: You can call SNPs, indels & SVs with high-performance variant calling nanopore data. Use #nanopore specific tools for best performance. Check out #EPI2ME Labs for workflows, tutorials & datasets: labs.epi2me.io 

SR: Remora – Oxford Nanopore’s latest methylation analysis tool - further enhances base-modification analysis on nanopore devices. Remora will be integrated into MinKNOW next week, enabling much simpler access to methylation data in real time. #nanoporeconf 

SR: 5mC, 5hmC, 6mA etc… are directly observable in the nanopore signal. #Nanopore is now the most accurate technology for characterising CpG #methylation. #nanoporeconf
SR: #Nanopore chemistry will read any fragment length. The read length lower limit is configurable.
SR: Short Fragment Mode (SFM) is designed to allow reads as short as 20 bases. @nanopore has demonstrated over 250M native human reads, with an average read length of ~200 bases, on a #PromethION Flow Cell: nanoporetech.com/about-us/news/… #nanoporeconf
SR: A new sequencing file format, .pod5, is designed to replace .fast5 and enable faster file writing. This supports increasing device outputs and accuracy, enabling smaller raw data file sizes and streamlining downstream analysis #nanoporeconf 

SR: Dorado is a new basecalling framework that will ultimately speed up access for users. Dorado is designed with support for @Apple GPUs & new @nvidia hardware & projected to keep up with high accuracy (HAC) models on #PromethION 48 #nanoporeconf
SR: A summary of our accuracies. #nanoporeconf 

Next up, @RosemaryDokos (RD) with a product update. 

RD: the #nanopore platform is fully scalable, offering the smallest flow cells with #Flongle and ultra-high throughput devices with #PromethION. Pricing of starter packs and flow cells is fully transparent on our web page: nanoporetech.com/products #nanoporeconf
RD: the platform is very versatile, and users can choose the best fit for their experiment; the answer to “how much data will I get?” often depends on which levers you vary #nanoporeconf 

RD: We apply the same principles to product release as to innovation; initial engagement with developers and early access users evolves into fully released products, allowing us to move fast and continuously improve #nanoporeconf 

RD: Short Fragment Eliminator Kit: Depletes short DNA fragments from your sample. Ultra-long DNA Sequencing Kit: ULK002, a move to Q20+ chemistry (Kit 14) kits with developers now. #nanoporeconf
RD: PCR cDNA Sequencing Kit: new adapter design increases flow cell output #PromethION > 160M reads & #MinION > 20M reads. #nanoporeconf
RD: Updates to #RNA sequencing. Single Cell Protocol: generate full-length transcripts from single cells, at high data volumes (>160M on #PromethION) enabling expression analysis. Direct #RNA protocol upgrade released into the community.
RD: . Critical work in public health contributing >1 million #SARSCoV2 genomes from 85 countries. The nanopore community made, and is still making, a significant contribution when the world needed it most. #nanoporeconf
RD: We have seen a strong community drive for broadening surveillance applications. This includes workflows for African Swine Fever, TB, respiratory metagenomics & routine pathogens such as #COVID19 and influenzas #nanoporeconf
RD: VolTRAX for #SARSCoV2 sequencing library prep - 4 sample per cartridge. #nanoporeconf
RD: We are preparing to launch Kit 14 versions of Ligation Sequencing Kit, the Native Barcoding Kit: 24 and 96, Cas9 barcoding method to early access customers and Rapid Barcoding Kit. #nanoporeconf 

RD: Stu's talk in one slide: key sequencing releases #nanoporeconf 

RD: The community are running high impact human projects: at scale, at speed and at depth. #nanoporeconf 

RD: #nanopore sequencing provides biology in full colour 

And now @The__Taybor (CB) to close with a few new and notable updates #nanoporeconf
CB: The MinION Mk1D. This is designed to be a tablet + keyboard with an integrated sequencer, VolTRAX too I hope, and will mean that users can simply dock a recommended tablet into the MinION Mk1D, pair it, and initiate sequencing. Register your interest: register.nanoporetech.com/minion-mk1d
CB: Nanopore sequencing is setting a new gold standard for methylation calling. #nanoporeconf
CB: Remora 1.0 models improve signal scaling which results in higher detection accuracy and quality filtered calls achieving 99.8% accuracy for 5mC in CpG contexts. #nanoporeconf
CB: Outy sequencing has potential to enable adaptive sampling + adaptive accuracy, & potential future application for highly accurate sequencing of rare molecules, single-molecule re-reading & variant calling. To find that needle in a haystack. #nanoporeconf
CB: Following analysis of Clive’s own genome and cell-free DNA isolated from plasma — the ‘cf Cliveome’ — using SFM, the team uncovered interesting biology, including plausible tissue typing from methylation data and greater insight into nucleosome arrangements #nanoporeconf 

CB: looking forward to raw sample to sequence - true liquid biopsy that could be deployed, for example, in a nanopore toothbrush. From end to end we can start with swab/spit, proof of concept now achieved #nanoporeconf 

This is designed for future environments where they don't have vortexes or pipettes. The sample could be wound pus, or a supermarket shelf #nanoporeconf
CB: What does the ideal device look like for decentralised sequencing? early blood glucose, 20 years ago, involved going to an office and giving blood. I think the ideal device would be a toothbrush - 50k white blood cells per ml in saliva. #nanoporeconf
CB: if we see something that looks suspicious, we can sequence it again and again and again. The toothbrush is on Wifi. Bonkers, never been done. This is where I want to take this technology. "Clive's fantasy product" #nanoporeconf
CB: There are lots of ways to sequence proteins on nanopores - some of which have been published. We think the same @nanopore platform that can be used to sequence DNA and RNA can be used for proteins #nanoporeconf 

CB: no. of steps a motor takes on DNA is >than one per amino acid. We put a construct on the pore, it looks like sequencing but we let the peptide go through the construction. We see a DNA signal 1st, then peptide signal-that was a struggle to see but now we can. #nanoporeconf
CB: as you can see here we are starting to generate profiles for more than one amino acid. They are very reproducible. #nanoporeconf 
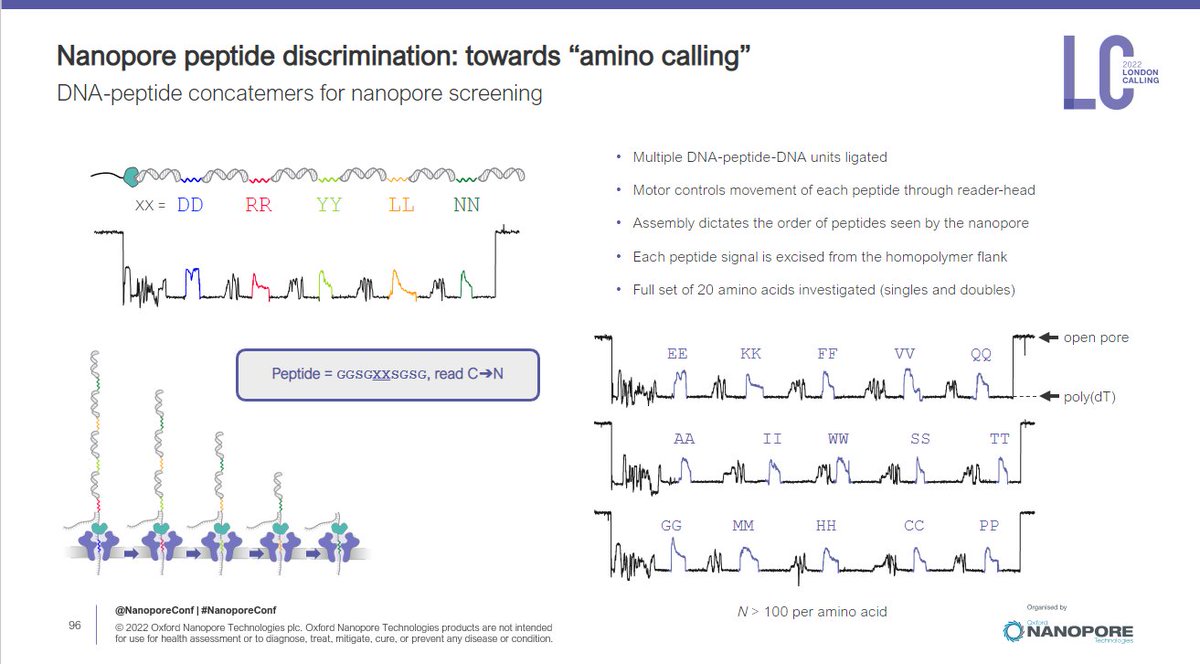
CB: we're learning a lot from this. #nanoporeconf 

CB: we want to start with a complex mix of proteins - a complex sample like blood, yeast, bacteria, and run on the same hardware that we use for DNA. So anyone could do complex proteomics on a MinION, GridION, PromethION. This is hard, very difficult #nanoporeconf 

CB: in summary, we can generate reproducible profiles of more than one amino acid - early proof of concept towards future protein sequencing #nanoporeconf
CB: in summary: we want to go from sample to answer easily, and to truly democratise liquid biopsy #nanoporeconf
@threadreaderapp thanks for unroll for this thread
• • •
Missing some Tweet in this thread? You can try to
force a refresh









