
🔴The curious case of Indian #Monkeypox Genomes
Wonderful effort by clinicians in @KeralaHealth who diagnosed the disease and researchers at @icmr_niv, we now have sequences of MPX isolates in @GISAID .
This short 🧵is on what the early genomes say.
Analysis @bani_jolly
Wonderful effort by clinicians in @KeralaHealth who diagnosed the disease and researchers at @icmr_niv, we now have sequences of MPX isolates in @GISAID .
This short 🧵is on what the early genomes say.
Analysis @bani_jolly

Briefly 4⃣ genome sequences have been deposited for two samples (EPI_ISL_13953610 and EPI_ISL_13953611) along with 2 re-sequenced genomes from isolates of one of the sample.
▶️Both the isolates were from early cases reported from Kerala
▶️Both cases have a travel history
▶️Both the isolates were from early cases reported from Kerala
▶️Both cases have a travel history
The present sustained human-human transmission of the MPX virus is believed to have happened via superspreader events in Europe with 16000+ cases now spread across 70+ countries,
The initial cases seemingly were predominantly among gays/bisexuals and msm networks
The initial cases seemingly were predominantly among gays/bisexuals and msm networks
A detailed publication on the initial 528 infections from 16 countries is summarised in the recent paper in @NEJM nejm.org/doi/full/10.10…
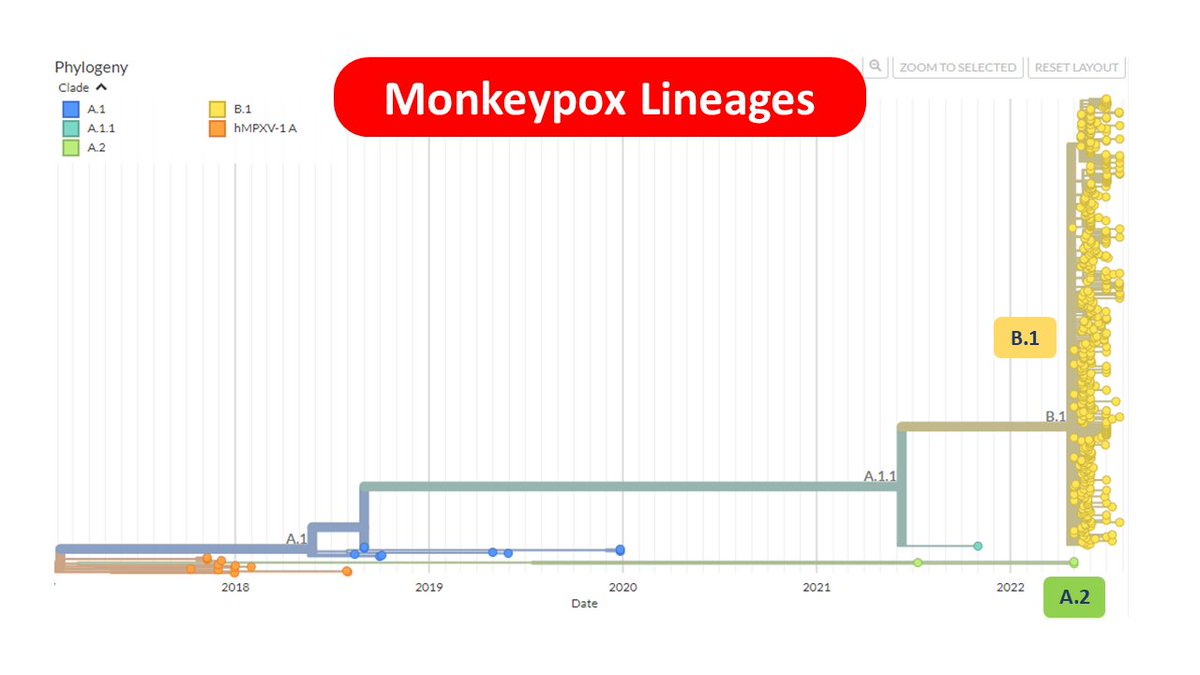
Many geomes from across the world sort of support this observation. This largely is represented as the B.1 lineage of the virus and encompass the predominant lineage for genomes in 2022.
However a very small no of genomes belong to a distinct cluster A.2
However a very small no of genomes belong to a distinct cluster A.2
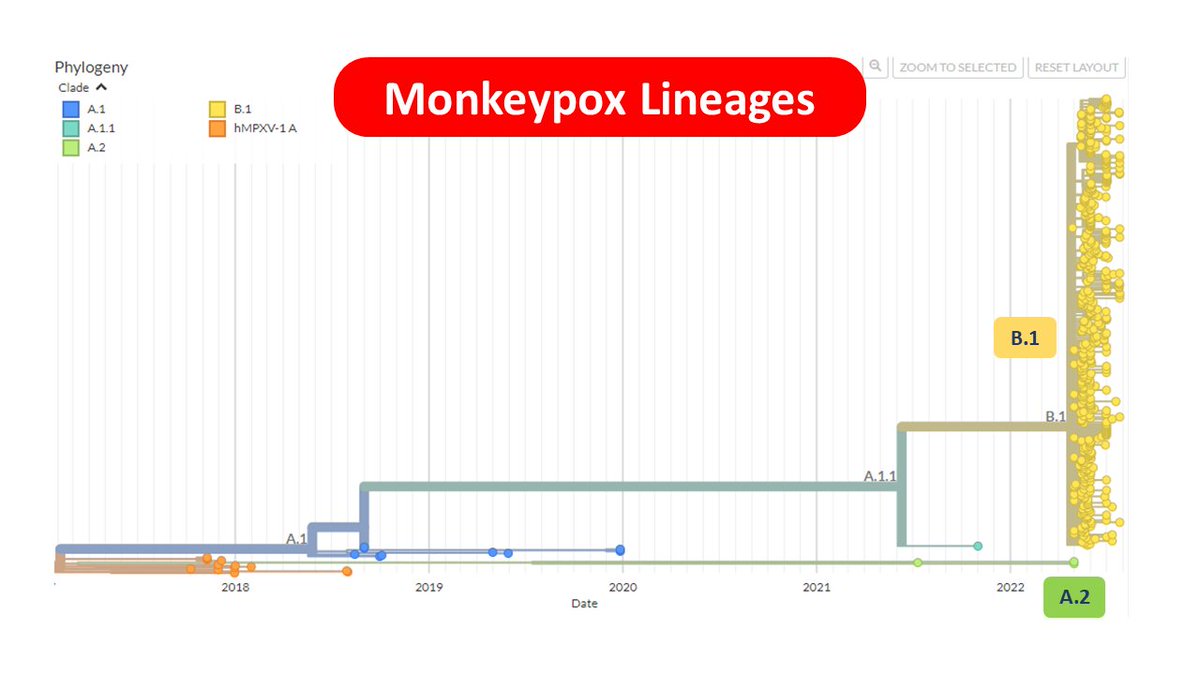
The A.2 cluster is peculiar in many aspects.
1⃣ Only a very few genomes belong to this cluster
2⃣The earliest genomes are from 2021
3⃣The genomes are from USA (Florida, Texas & Virginia) and Thailand
4⃣Cluster does not show polytomy suggestive of a superspreader event.
1⃣ Only a very few genomes belong to this cluster
2⃣The earliest genomes are from 2021
3⃣The genomes are from USA (Florida, Texas & Virginia) and Thailand
4⃣Cluster does not show polytomy suggestive of a superspreader event.
There is a detailed article on this distinct cluster by @HelenBranswell
https://twitter.com/HelenBranswell/status/1532751642977869825?s=20&t=gLKjhbZxQgDLqqIPCjHj0A
Incidentally the two genomes from #Kerala map to this small distinct cluster A.2
This would mean
1⃣ These cases are not possibly linked to the european superspreader events.
2⃣ We might be looking at a distinct cluster of human-human transmission and possibly unrecognised for yrs
This would mean
1⃣ These cases are not possibly linked to the european superspreader events.
2⃣ We might be looking at a distinct cluster of human-human transmission and possibly unrecognised for yrs
Incidentally the two cases from Kerala have a travel history to the Middle East, and so do some other cases in this cluster have travel history to Middle East / East Africa
The tMRCA for the cluster (time to most recent common ancestor) is mid 2021 (caveat-small no of samples).
The earliest sample in the cluster from USA is indeed from 2021 suggesting the virus has been in circulation for quite some time, and earlier than the European events.
The earliest sample in the cluster from USA is indeed from 2021 suggesting the virus has been in circulation for quite some time, and earlier than the European events.
In conclusion
1⃣ Kerala MPX genomes belong to A.2 in contrast to majority of the genomes across the world which belongs to B.1 lineage
2⃣ Genomes can uncover many interesting facts & leads. Epidemiologists need to follow up leads
3⃣ Genome surveillance is invaluable in outbreaks
1⃣ Kerala MPX genomes belong to A.2 in contrast to majority of the genomes across the world which belongs to B.1 lineage
2⃣ Genomes can uncover many interesting facts & leads. Epidemiologists need to follow up leads
3⃣ Genome surveillance is invaluable in outbreaks
Taking forward,
🔹We should be looking forward to more genomes as cases emerge and possibly sequence every single case.
🔹Public Health measures and communication needs to take these new insights into consideration
🔹Wide testing and awareness could uncover many more cases.
🔹We should be looking forward to more genomes as cases emerge and possibly sequence every single case.
🔹Public Health measures and communication needs to take these new insights into consideration
🔹Wide testing and awareness could uncover many more cases.
To reiterate facts, and give credits where it is due, the sequences were deposited by @icmr_niv in @GISAID and the cases were worked up by clinicians in @KeralaHealth
• • •
Missing some Tweet in this thread? You can try to
force a refresh









