
#KRAS alert! Over the past 4 years, a multi-disciplinary team from @hopkinskimmel @VUMC_Cancer @DanaFarber @MSKCancerCenter & @Amgen has been studying the distribution and co-mutations of KRAS, NRAS and HRAS mutant #cancers under the auspices of @AACR project #GENIE. Read the 🧵 

Our work is now published online at @CR_AACR and represents the largest study of RAS genomic architecture to date, leveraging #NGS data from >600,000 mutations and >66,000 tumors across 51 #cancer types.
Full text here
aacrjournals.org/cancerres/arti….
Full text here
aacrjournals.org/cancerres/arti….
Our findings provide insights in the genomic architecture of RAS mutant cancers and may serve as a blueprint for mapping therapeutic vulnerabilities for mut RAS. Given the vast array of findings we have put together a shiny app for interactive #dataviz:
ras-hallmarks.jhmi.edu
ras-hallmarks.jhmi.edu
We studied the prevalence and genomic diversity of RAS in human cancers with a mutant allele resolution and considered host contexts determined by age, self-reported race and sex & tumor backgrounds determined by cell lineage, TMB and mutational signatures. Context matters.
We estimated the cancer type-specific prevalence of KRAS, NRAS, and HRAS mutant alleles at codons 12, 13 & 61 using a Bayesian hierarchical model by age, race and sex. This approach allows information to be shared across cancer types and strata through the hierarchical structure. 

We then assessed co-mutation patterns with non-RAS mutations, copy number alterations, and rearrangements using a Bayesian hierarchical model. This allows for information on co-occurrence to be shared across cancer types and shrunk estimates for rarer cancer types.
These analyses showed a cancer lineage-dependent distribution of RAS isoform prevalence that differed by sex, age and racial background. The context-dependent distribution of mutant RAS should be considered in target population selection & rational #clinicaltrial design. 

RAS mutations co-occurred with mut ATM, KEAP1, MAX, NKX2-1, RBM10, STK11 & U2AF1 in #lungcancer, CDKN2A in #PancreaticCancer, PIK3CA in colon cancer, ARID1A & PTEN in uterine cancer and TERT in #Melanoma. RAS/TP53 co-mutations were less frequent than expected in several cancers. 

RAS/RAS co-mutations were infrequent. Notably, we found RAS/RAS co-muts in 4.4% of ovarian cancers, 4.9% of uterine cancers & 11.7% of bladder cancers. HRAS/NRAS co-muts were found in bladder cancer (5.9%), while KRAS/NRAS & KRAS/KRAS co-muts were found in ovarian cancer (1.8%).
Differential co-mut patterns were seen at the mutant allele level; as an example KRAS G12C co-occurred with ATM, RBM10, KEAP1, NTRK3, EPHA5, AMER1 and STK11 mutations in NSCLC. RBM10/KRAS G12A & G12D co-muts and KRAS G12D/NKX2-1 & MAX co-muts were also detected in #lungcancer. 

Looking at convergence of co-muts in signaling pathways, we found mutual exclusivity of nonRAS muts in the RAS/MAPK pathway with KRAS G12A, G12C, G12D and less with G12V in NSCLC, NRAS Q61K & Q61R in melanoma and NRAS Q61R in thyroid cancer; but not in colon or pancreatic cancers 

Mutations in DDR genes, chromatin regulating genes and genes in the NRF2/oxidative stress response pathway were enriched in KRAS G12C-mutant NSCLC. A different co-mut pattern was noted for CRC, where KRAS G12D and G13D we found to be co-mutated with genes in the PI3K/AKT pathway.
Triple co-mutations followed cancer cell lineage specific distributions. Notably a number of triple mutations were detected in CUP, including KRAS/KEAP1/STK11, KRAS/KEAP1/SMARC4, KRAS/KEAP1/TP53, KRAS/STK11/SMARC4, KRAS/KEAP1/PTPRD, KRAS/PTPRD/TP53 and KRAS/STK11/TP53.
❗️❗️Co-mutation patterns can be confounded by accumulating passenger mutations and mutational signatures that can be differentially susceptible to mutation; co-occurrence of genomic alterations may thus reflect TMB/mutational spectra rather than positive selection.
The strength of specific RAS dependencies was attenuated in different TMB backgrounds. Most susceptible co-muts: KRAS/STK11, KEAP1 or TP53 mutations in NSCLC and TP53 and inactivating mutations in APC in CRC. KRAS G12C/STK11 & KRAS G12C/KEAP1 mutations enriched in low TMB. 

KRAS G12C/KEAP1 or STK11 co-muts were less frequent in tumors with a smoking or an APOBEC mut signature, while KRAS G12C mut NSCLC with an APOBEC mut signature more frequently harbored NTRK3 mutations. In CRC, KRAS/ PIK3CA co-occurred in tumors with an APOBEC mut signature.
Next step was to investigate differential transcriptomic tracks in tumors with differential co-muts, with respect to cancer hallmarks as well as immune surveillance states. We found discrete transcriptional programs enriched in RAS mut tumors in a cancer lineage-dependent manner. 
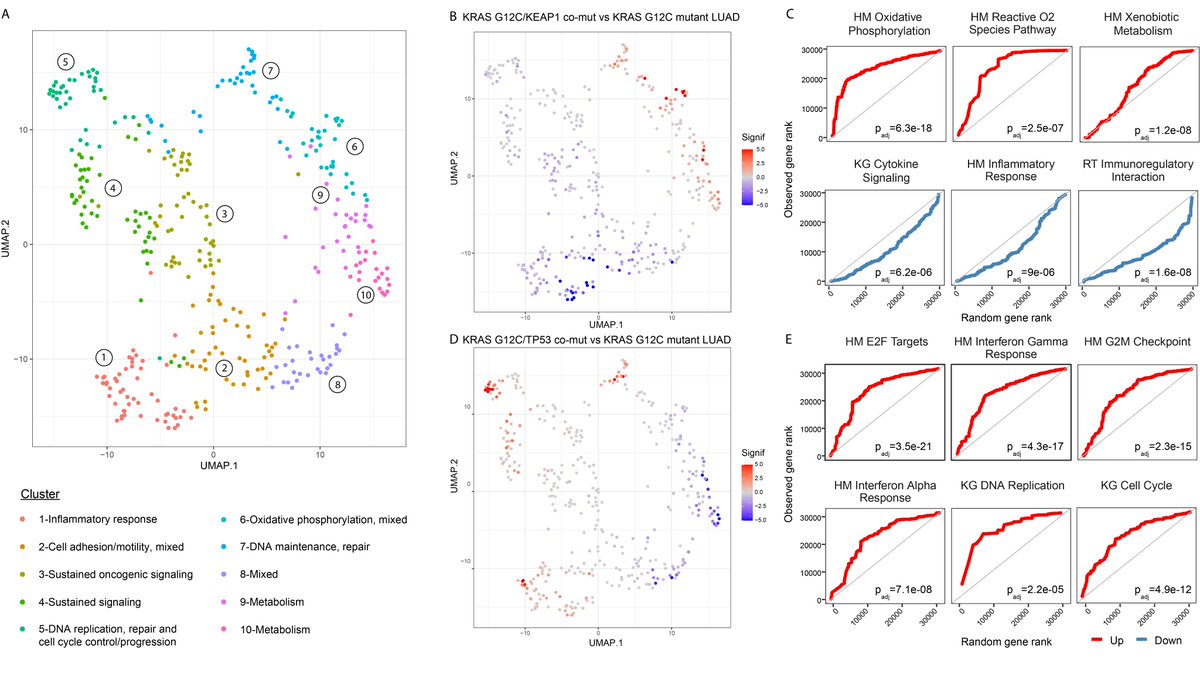
We hypothesized that these distinct phenotypes of KRAS mutant tumors may reflect clinical outcomes and therapeutic vulnerabilities and evaluated the association between the distinct genomic tracks of RAS mutant tumors with clinical outcomes.
In addition to TCGA co-mut stratified survival analyses, we confirmed that KRAS G12C/STK11 co-muts conferred shorted overall survival with immunotherapy. In contrast, patients with NSCLC harboring KRAS G12C and TP53 co-muts attained longer overall survival with immunotherapy. 

Overall, our study assessed the genomic landscape of RAS mutant tumors in a pan-cancer manner and with a RAS mutant allele-specific resolution, considering gene-gene interactions individually as well within gene families and pathways and accounting for host and tumor confounders.
This work is a testament to collaborative research, bringing together a phenomenal team: Rob Scharpf Archana Balan @BiagioMd Chris Cherry CG Wang Michele Newton Hira Rizvi James White Alex Baras @Blair_Landon15 Jocelyn Lee Hil Hsu @RielyMD @DrMarkAwad @christine_lovly
Here’s the @MolecularOncLab computational team seeing the paper go live! Archana Balan @Blair_Landon15 James White @Marta_M_Agrawal @PaolaGhanem Chris Cherry @HopkinsThoracic @CMM_JHU @JHSPHBMB @OslerResidency congrats on your phenomenal work! Until the next one 😉! 

• • •
Missing some Tweet in this thread? You can try to
force a refresh



