Reflecting on today's fantastic #iMED2022, I am thrilled to share an app I've been creating to create high-quality🤓, collaborative🗺️, & dynamic 📈diagnostic #MedEd schemas!
Schematify App Here: diagnostic-schema.herokuapp.com
Video:
Check it out: 1/
Schematify App Here: diagnostic-schema.herokuapp.com
Video:
Check it out: 1/
Medical learning is all about the organization of a HUGE amount of (growing) knowledge😅
I loved the concept of a logical framework...but my handwriting, spelling and ability to hold onto paper documents is👎🙈
Plus, I wanted to be able to collaborate dynamically with YOU!🤩 2/
I loved the concept of a logical framework...but my handwriting, spelling and ability to hold onto paper documents is👎🙈
Plus, I wanted to be able to collaborate dynamically with YOU!🤩 2/
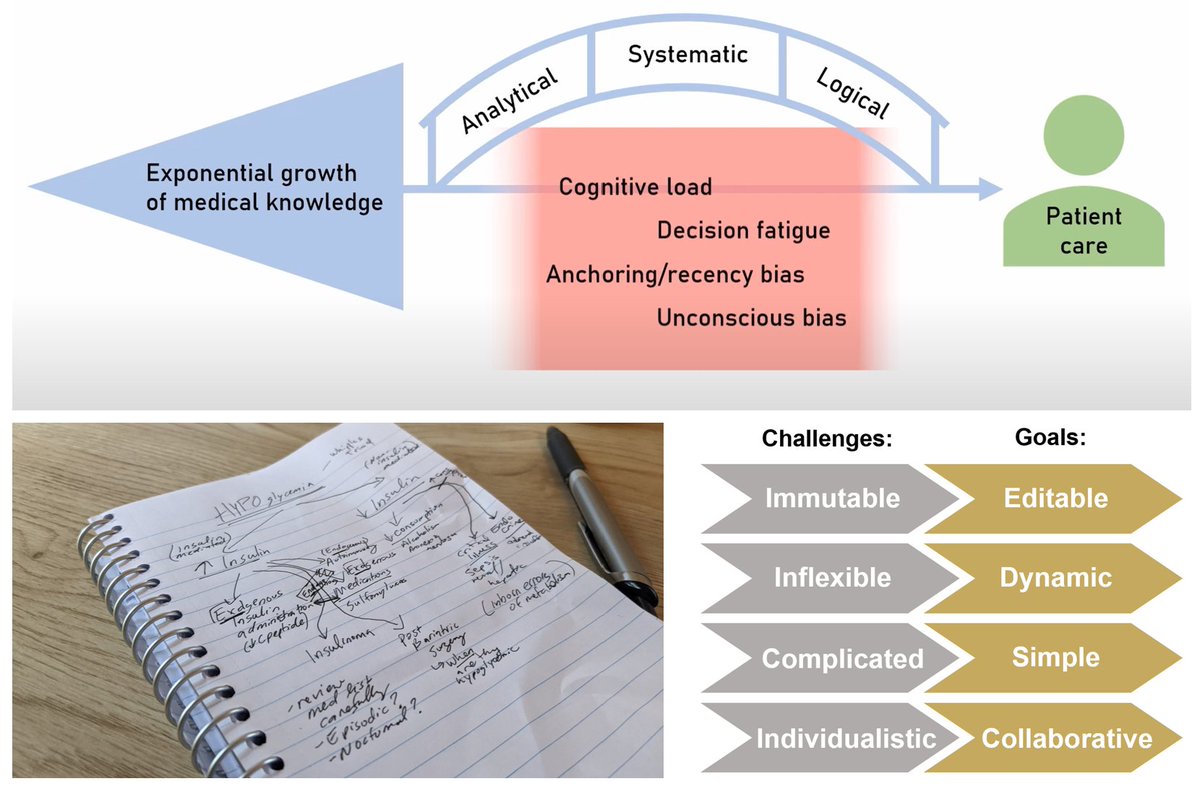
Current tools (handwritten, PowerPoint, chart apps) all had their limitations and were not designed for medical trainees!
So I used @ThePSF #python 🐍with @plotlygraphs Dash and @Graphviz to develop a web tool that leverages the collaboration capabilities of @googlesheets 3/
So I used @ThePSF #python 🐍with @plotlygraphs Dash and @Graphviz to develop a web tool that leverages the collaboration capabilities of @googlesheets 3/
Enter your content in the sheet, make it public, share with friends, and copy the URL into the app for instant simple schemas that you can export as PDFs 🤯
Sheet Template: docs.google.com/spreadsheets/d…
And adding a new node or branch is as simple as adding a row to your sheet!🙌4/
Sheet Template: docs.google.com/spreadsheets/d…
And adding a new node or branch is as simple as adding a row to your sheet!🙌4/

You may also be able to envision new ways to integrate this into #MedEd teaching.💡
For example, the Vandy Problem Solvers @VUmedicine @VUMCMedicineRes with leadership from @chasejwebber allowed me to create a schema in real-time while we worked through a case conference🕷️5/
For example, the Vandy Problem Solvers @VUmedicine @VUMCMedicineRes with leadership from @chasejwebber allowed me to create a schema in real-time while we worked through a case conference🕷️5/
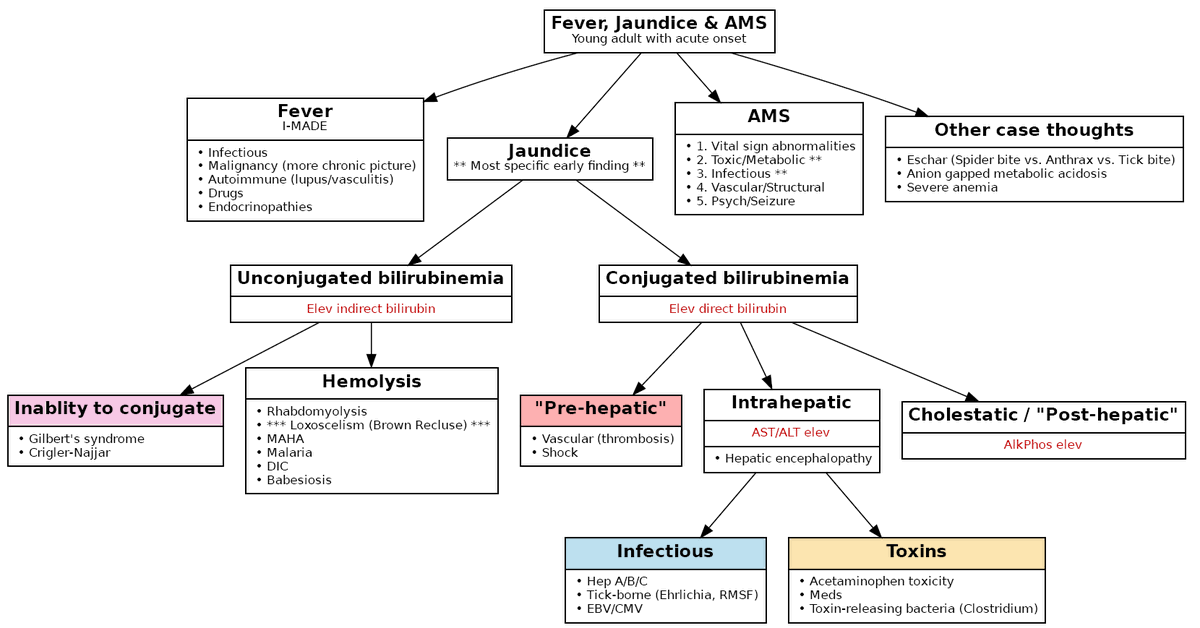
This is still a work in progress (you will find bugs🐞) but I hope it might be helpful!
Thanks to so many that inspired me with their schemas, diagnostic thinking, digital tools, and discussion especially @chasejwebber
Code: github.com/emcarthur/diag…
Please reach out! 6/6 ✉️
Thanks to so many that inspired me with their schemas, diagnostic thinking, digital tools, and discussion especially @chasejwebber
Code: github.com/emcarthur/diag…
Please reach out! 6/6 ✉️

• • •
Missing some Tweet in this thread? You can try to
force a refresh









