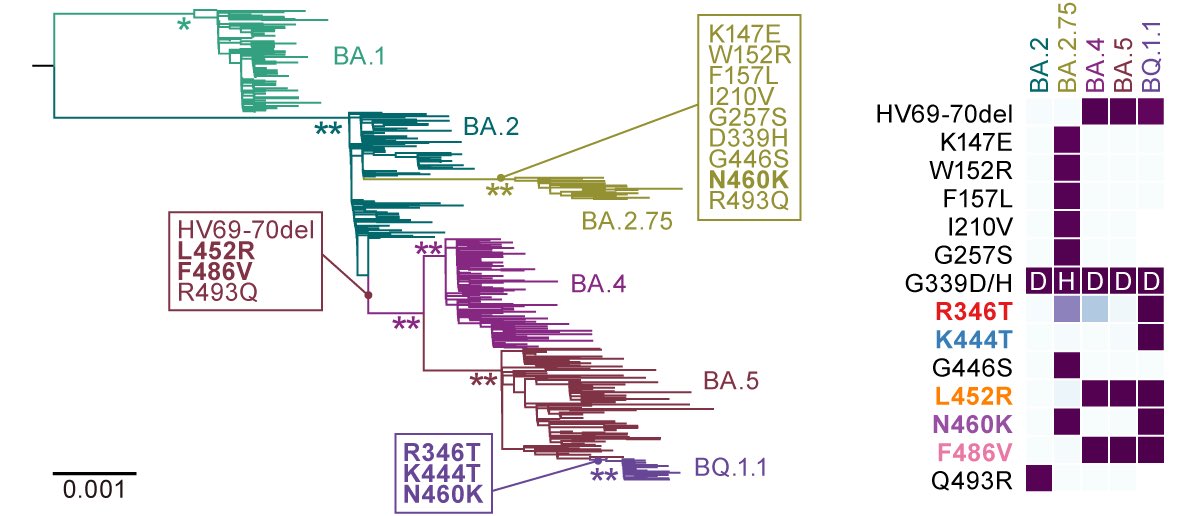
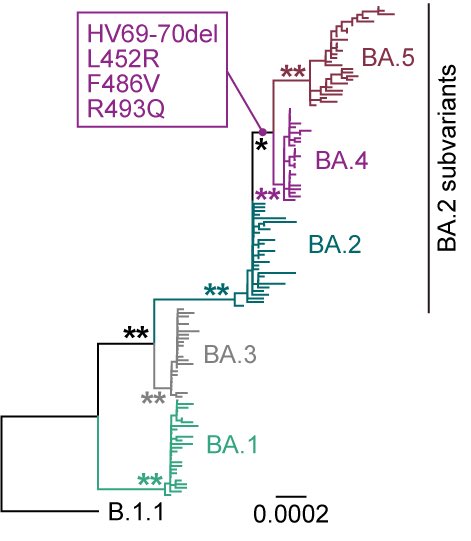
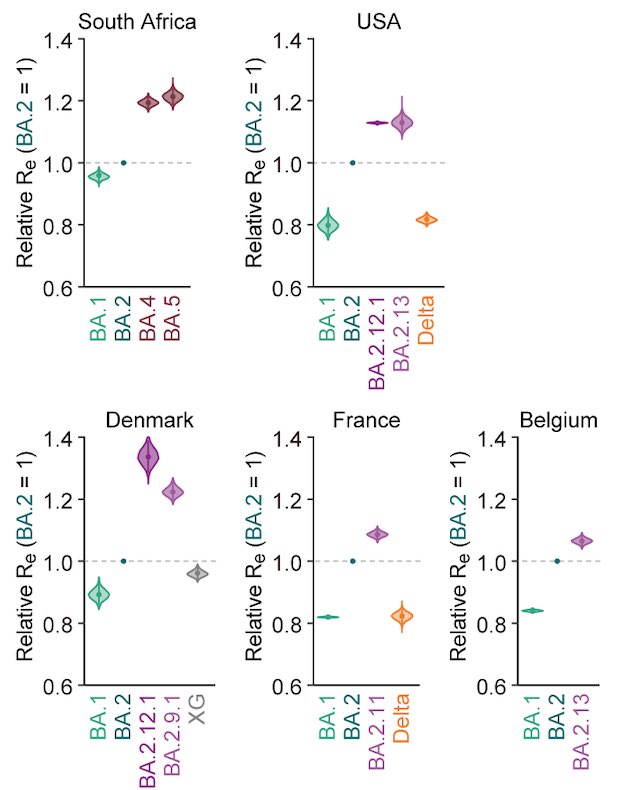
BREAKING🔔 The 15th paper from G2P-Japan🇯🇵 is out at iScience @iScience_CP. We revealed that the virological features of #Omicron BA.1 variant are determined by the spike #S375F mutation. Please RT. 1/6
cell.com/iscience/fullt…
cell.com/iscience/fullt…
We have demonstrated that the Omicron BA.1 spike exhibits 1) decreased efficiency of spike cleavage and 2) attenuated fusogenicity:
Cf. The 5th paper from G2P-Japan🇯🇵↓
nature.com/articles/s4158…
However, it remained unclear which mutation(s) determine these BA.1’s features. 2/6
Cf. The 5th paper from G2P-Japan🇯🇵↓
nature.com/articles/s4158…
However, it remained unclear which mutation(s) determine these BA.1’s features. 2/6
1️⃣Our comprehensive screening experiments revealed that these BA.1's features are determined by its receptor binding domain and particularly, only #S375F mutation is responsible. 3/6
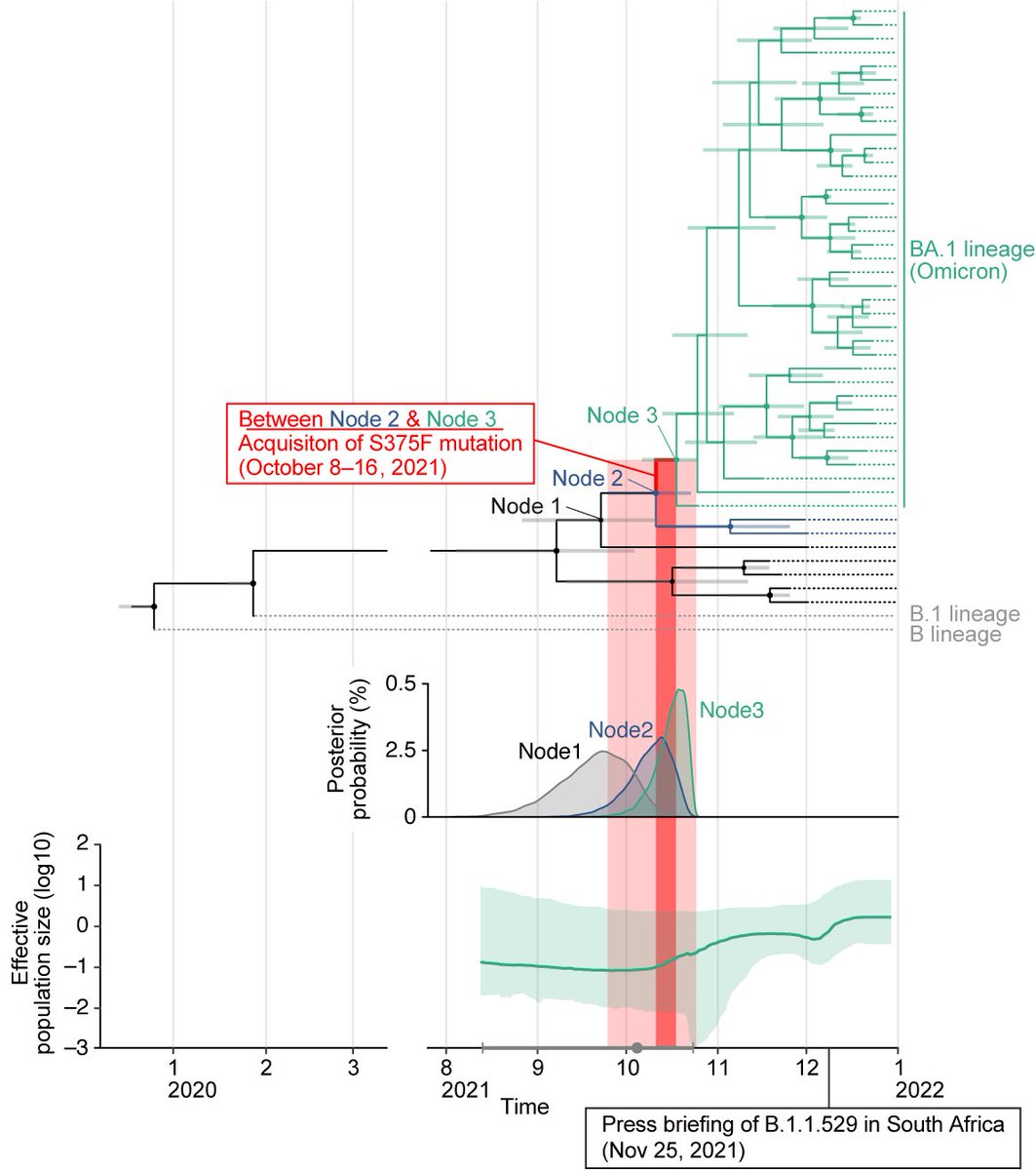
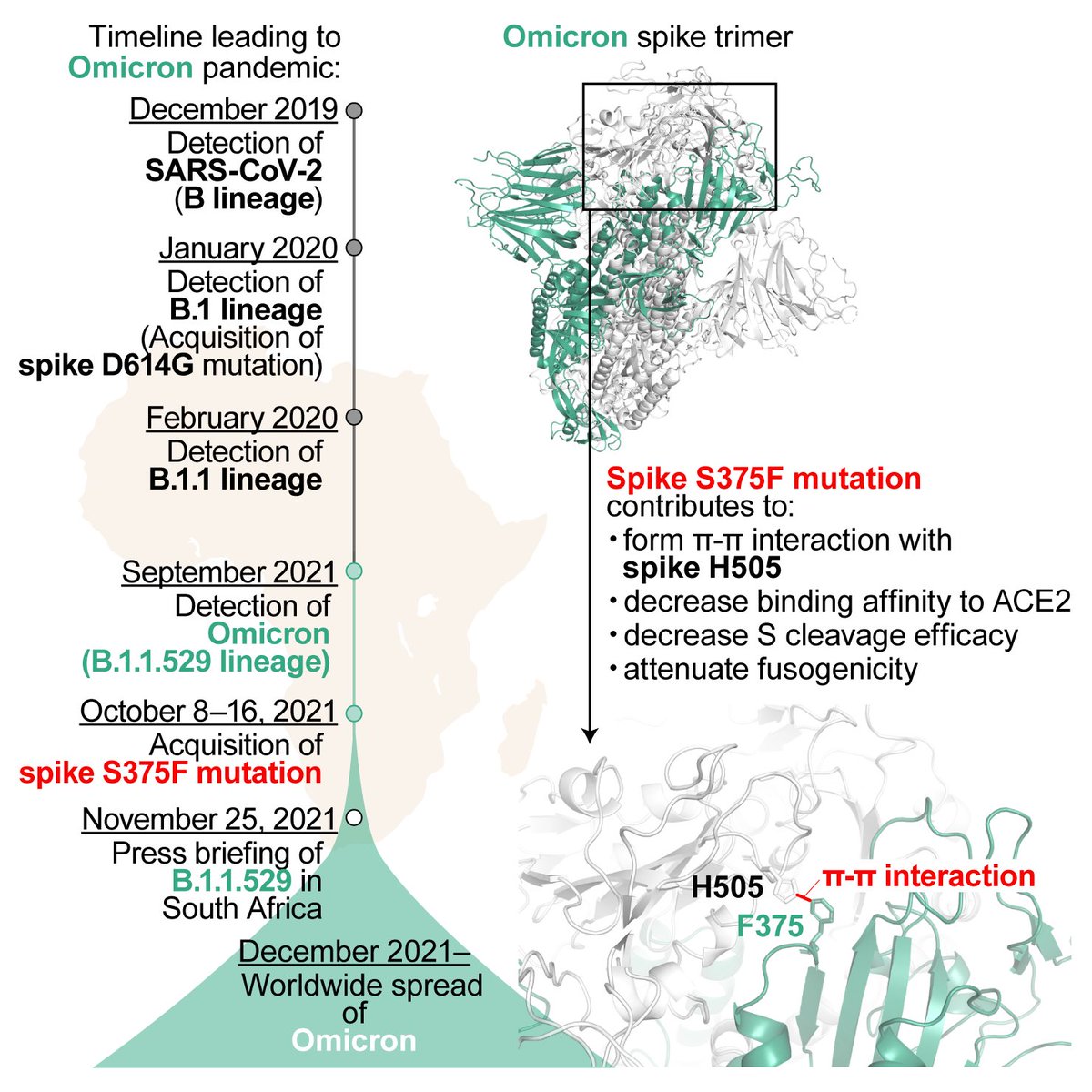
2️⃣Interestingly, molecular phylogenetic analysis showed that #S375F mutation was acquired by ancestral Omicron just before the explosive spread of this variant in South Africa🇿🇦, suggesting that the acquisition of S375F mutation was a trigger of the massive spread of Omicron. 4/6 
3️⃣In-depth structural analysis showed that the F375 residue in a spike protomer can form inter-protomer pi-pi interaction with the H505 residue in another spike protomer, which confers attenuated spike cleavage efficacy and fusogenicity. 5/6 
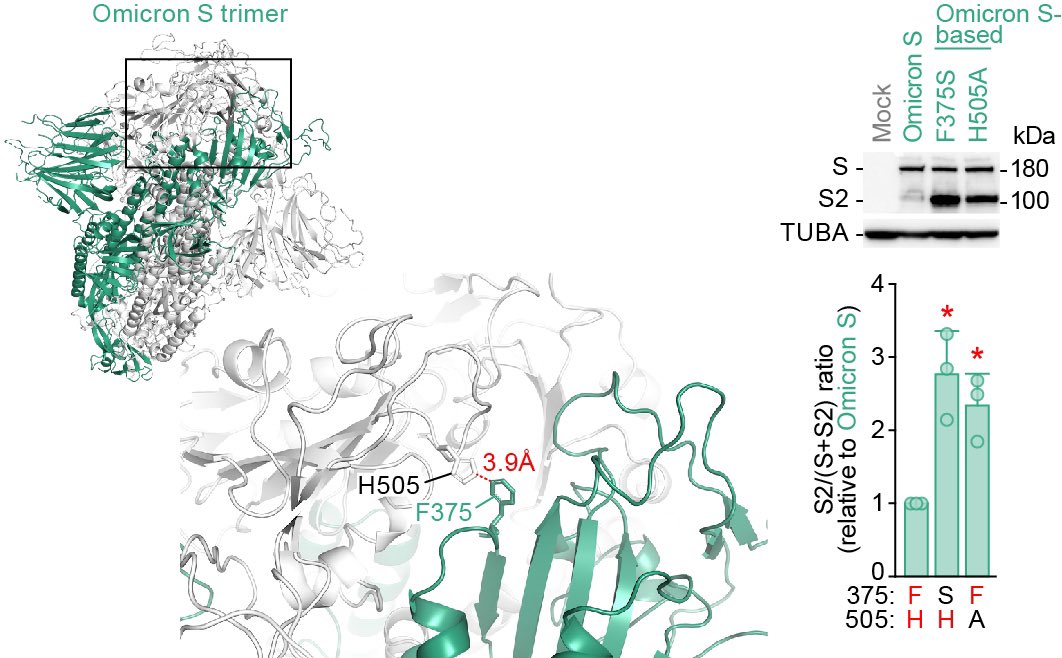
▶️This is a graphical summary of this study. Our data shed light on the evolutionary events underlying Omicron emergence at the molecular level. These are the results of multi-scale interdisciplinary investigations, THE feature of G2P-Japan🇯🇵. Hope you like it! 6/6 
FYI This is the tweet introducing original preprint in April 2022:
https://twitter.com/systemsvirology/status/1510928817535488004
• • •
Missing some Tweet in this thread? You can try to
force a refresh