BREAKING🔔 The 15th preprint from G2P-Japan🇯🇵 is out @biorxivpreprint. We illuminated the virological characteristics of one of the latest SARS-CoV-2 lineages of concern, XBB, which was generated by recombination of two #Omicron subvariants. Please RT. 1/
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
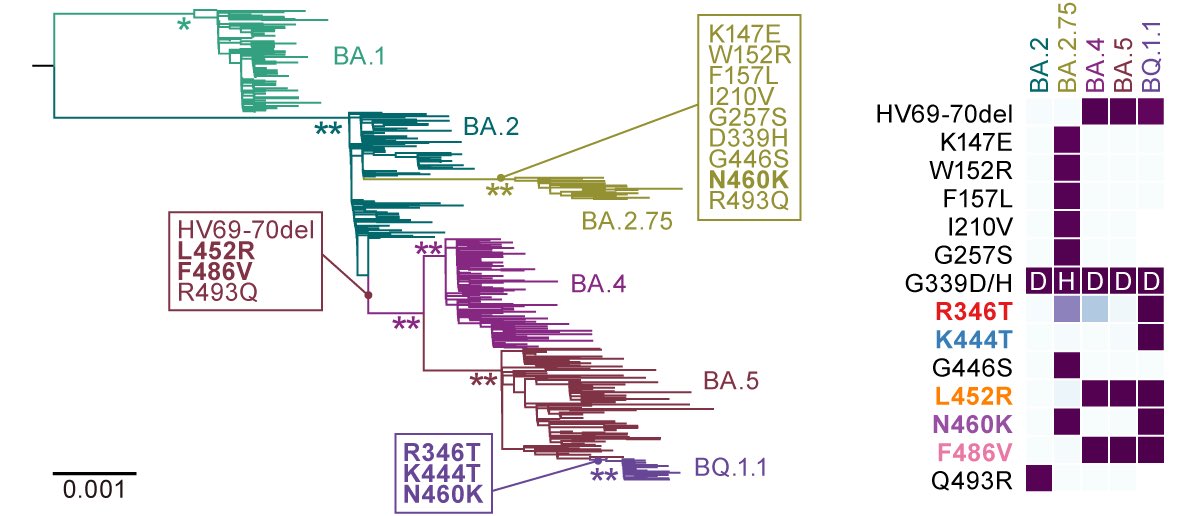
In late 2022, the Omicron subvariants have highly diversified. An example is BQ.1.1, which has convergently acquired amino acid substitutions at five critical residues in the spike protein: R346T, K444T, L452R, N460K & F486V. We reported it as below⬇️ 2/
https://twitter.com/SystemsVirology/status/1599909576442544128?s=20&t=y37dS78E_Po8bhJZUrRzEw
Another variant of concern is XBB (aka #gryphon). First, two young talents, @jampei2 & @SpyrosLytras, showed that XBB emerged by recombination of two co-circulating BA.2 lineages, BJ.1 & BM.1.1.1 (a progeny of BA.2.75), during 2022 summer in India or its neighboring countries. 3/ 

We showed that the Re values of XBB lineages are comparable with or slightly higher than those of BQ.1 lineages, and notably, XBB and BQ.1 lineages are becoming dominants in Eastern and Western Hemisphere, respectively. 4/ 

Neutralization assay revealed that XBB is the most profoundly resistant variant to BA.2/5 breakthrough infection sera ever (!!). >30-fold more resistant to BA.2 breakthrough infection sera than BA.2, and >13-fold more resistant to BA.5 breakthrough infection sera than BA.2😵 5/ 

XBB spike is more fusogenic than BA.2.75 spike. Notably, the recombination breakpoint is located in the receptor-binding domain of spike, and each region of recombined spike respectively conferred both immune evasion and augmented fusogenicity to the XBB spike. 6/ 

Finally, we demonstrated that the intrinsic pathogenicity of XBB in hamsters is comparable to or even lower than that of BA.2.75, an ancestral lineage of XBB. 7/ 



Our multiscale investigation provided evidence suggesting that #XBB is the first documented example of a SARS-CoV-2 variant that increased its fitness (Re) through an recombination event rather than substitutions. 8/8 

• • •
Missing some Tweet in this thread? You can try to
force a refresh