Hello!
📢A new paper in which I collaborated with the group of @Rocha_Lab is now available in @PNASNews !!!
📰pnas.org/doi/10.1073/pn…
1/13- I always thought that synthetic hydrogels were relatively limited in terms of how physiologically representative they are.
📢A new paper in which I collaborated with the group of @Rocha_Lab is now available in @PNASNews !!!
📰pnas.org/doi/10.1073/pn…
1/13- I always thought that synthetic hydrogels were relatively limited in terms of how physiologically representative they are.
2/13 - Synthetic hydrogels are typically linear elastic, which is cool, because it makes my life easier while doing Traction Force Microscopy (TFM) on them 🤓... but they don't display strain stiffening characteristics as they are non-fibrous.
3/13 - 🛠️Cells in natural hydrogels, like collagen, reorient fibers around them, and they can mechanically communicate with other cells that are at a certain distance from them by remodeling and stiffening the matrix in their surroundings
But it turns out that I was wrong...
But it turns out that I was wrong...
4/13 - Polyisocyanide (PIC) hydrogels actually display many of those features that are so relevant to mimic physiological conditions within in vitro models!
And you can still tune their mechanical properties very easily!
In this paper we showed some of those features:
And you can still tune their mechanical properties very easily!
In this paper we showed some of those features:
5/13 - First of all, we saw that cells remodel the matrix, densifying the peri-cellular region, very similarly as they do in collagen hydrogels.
🧐Look:
🧐Look:

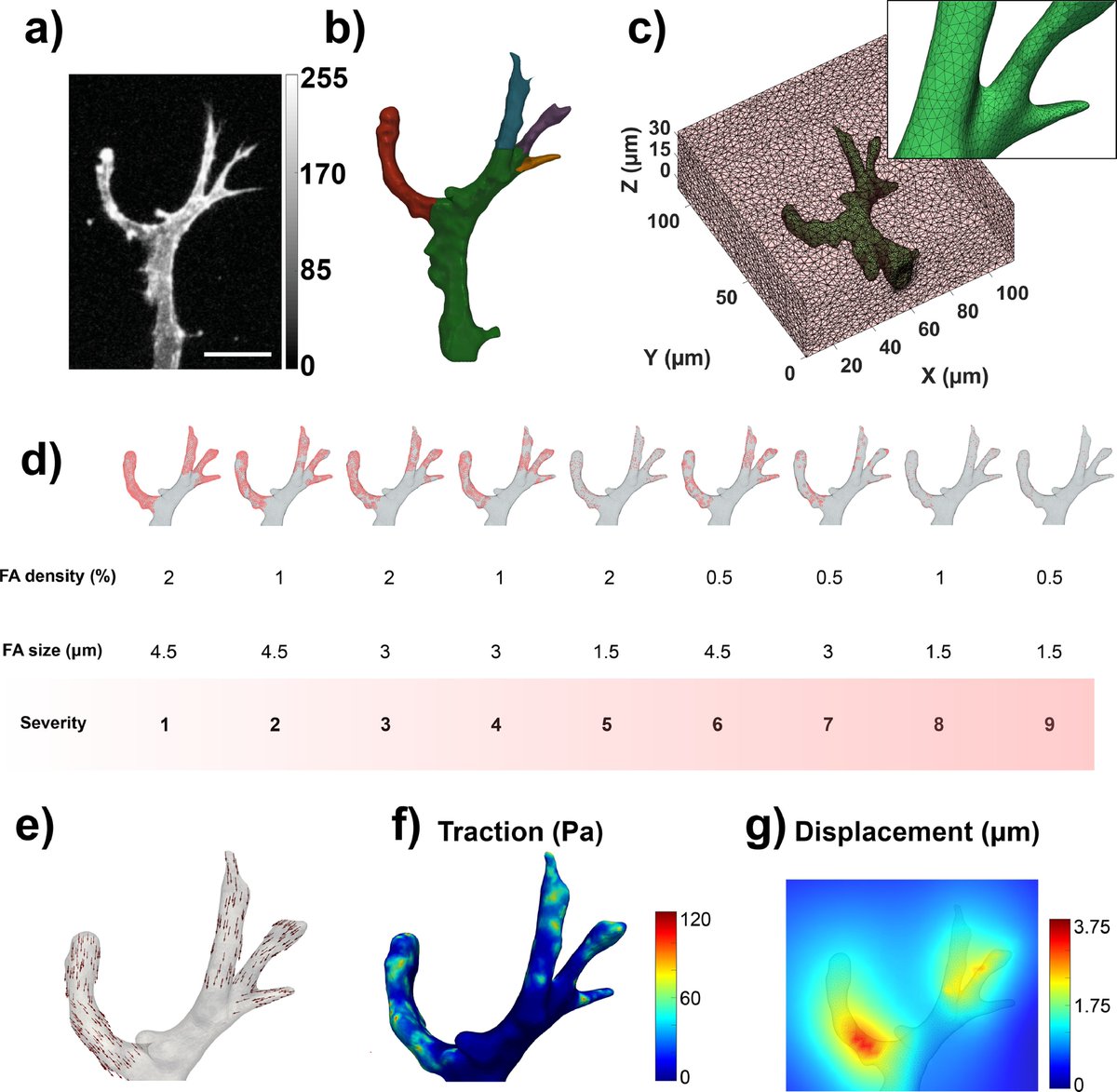
7/13 - Then, of course🤓, we used #TFMLAB to measure the 3D hydrogel deformations induced by cell forces💪💪
💚I am particularly very excited about these results because it is the first time that TFMLAB has been used in an external lab and it has proven to be quite handy!
🧐
💚I am particularly very excited about these results because it is the first time that TFMLAB has been used in an external lab and it has proven to be quite handy!
🧐

8/13 - Those displacement field patterns looked very much like what I typically see in collagen hydrogels, with slow decays as you move away form the cell.
But to be sure, we also showed displacement fields in collagen and matrigel:
But to be sure, we also showed displacement fields in collagen and matrigel:

9/13-We also quantified cell volume, sphericity, the number of cell protrusions, their length, and the differences in matrix displacements and we saw that there are barely any differences between PIC and collagen.
The way I understand this is: cells are equally happy in both 😍
The way I understand this is: cells are equally happy in both 😍

10/13 - Btw, you might wonder how did we quantify those parameters in 3D data.
We developed fully automated custom-codes for it.
If you need similar stuff, I still have the codes, just contact me😉
🧐Look at this of the automatic segmentation of cell protrusions:
We developed fully automated custom-codes for it.
If you need similar stuff, I still have the codes, just contact me😉
🧐Look at this of the automatic segmentation of cell protrusions:

11/13 - We also did an algorithm to segment cells in 3D and that are touching each other. It is sometimes tricky, but we wrote something based on watersheds that worked beautifully:
Image shows projections, but it works in 3D (which allows to quantify 3D morphological stuff)
Image shows projections, but it works in 3D (which allows to quantify 3D morphological stuff)

12/13 - A reminder that #TFMLAB is open source and that you can download the code here:
gitlab.kuleuven.be/MAtrix/Jorge/t…
gitlab.kuleuven.be/MAtrix/Jorge/t…
13/13 - I'm sharing also the nice thread that the main PI of the project, @Rocha_Lab , shared when the preprint was out. She explains more things there:
https://twitter.com/Rocha_Lab/status/1563193032224018433?s=20
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter