
Statistical geneticist @ Regeneron | PhD @broadinstitute & @harvardmed | tweets are my own
How to get URL link on X (Twitter) App

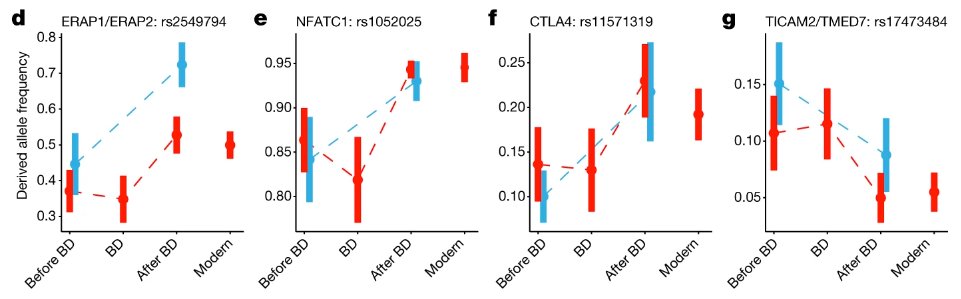
 It's so impressive they got 206 ancient DNA samples pre-, during, and post- plague in not 1, but 2 cities (so they could replicate the findings). I'm honestly gobsmacked at how cool this paper is. Huge congrats to Jennifer Klunk, @TaurVil, @LB_Barreiro and co.🎉
It's so impressive they got 206 ancient DNA samples pre-, during, and post- plague in not 1, but 2 cities (so they could replicate the findings). I'm honestly gobsmacked at how cool this paper is. Huge congrats to Jennifer Klunk, @TaurVil, @LB_Barreiro and co.🎉

 Across 454k individuals, they found 12.3M variants (99.6% MAF<1%). This is:
Across 454k individuals, they found 12.3M variants (99.6% MAF<1%). This is:

 Testing individual coding variants (so no burden tests), they found 975 associations (P<5e-8), 715 at Bonferroni threshold (P<2e-9). Most associations in missense variants (although unclear whether that is enriched above expectation), and 1/3 were novel.
Testing individual coding variants (so no burden tests), they found 975 associations (P<5e-8), 715 at Bonferroni threshold (P<2e-9). Most associations in missense variants (although unclear whether that is enriched above expectation), and 1/3 were novel.

 Within 3 cohorts with #COVID19 phenotypes (@uk_biobank, @GeisingerHealth, @PennMedicine), we analyzed rare variants (MAF<0.5%; ~7million) and burden tests within each ancestry separately and meta-analyzed our results. Incorporating non-EUR data increased our case N by ~10%.
Within 3 cohorts with #COVID19 phenotypes (@uk_biobank, @GeisingerHealth, @PennMedicine), we analyzed rare variants (MAF<0.5%; ~7million) and burden tests within each ancestry separately and meta-analyzed our results. Incorporating non-EUR data increased our case N by ~10%.
