Chief Scientific Officer, Nightingale Health. Formerly led the Covid-19 genomics initiative @sangerinstitute.
6 subscribers
How to get URL link on X (Twitter) App





https://twitter.com/jbloom_lab/status/1468001919566184448?s=20


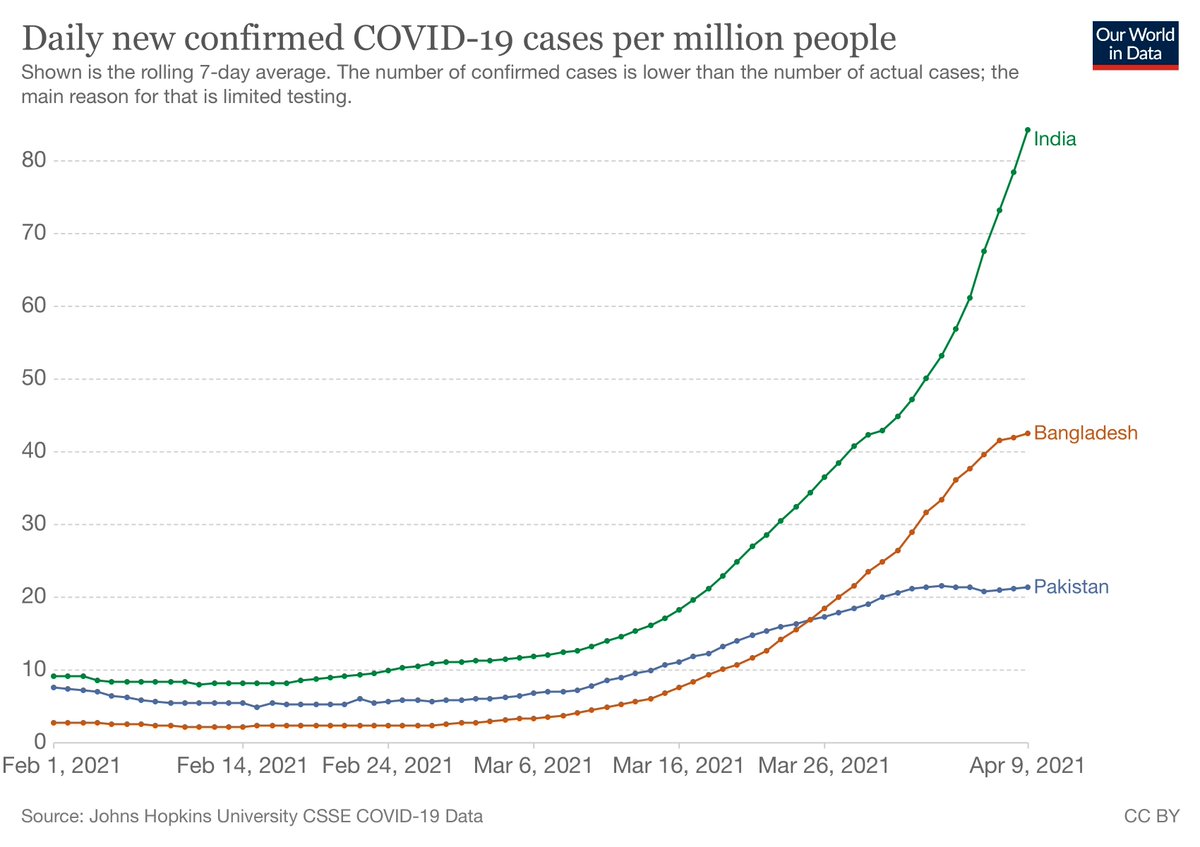
https://twitter.com/DataDrivenMD/status/1466501188858232834The thread looks at one mutation (del69/70) that's in Omicron, Alpha and some Delta in plots from outbreak.info. It asserts older sequences with del69/70 may have been "misclassified" because "we didn't know about Omicron". But we don't look at one mutation at a time!

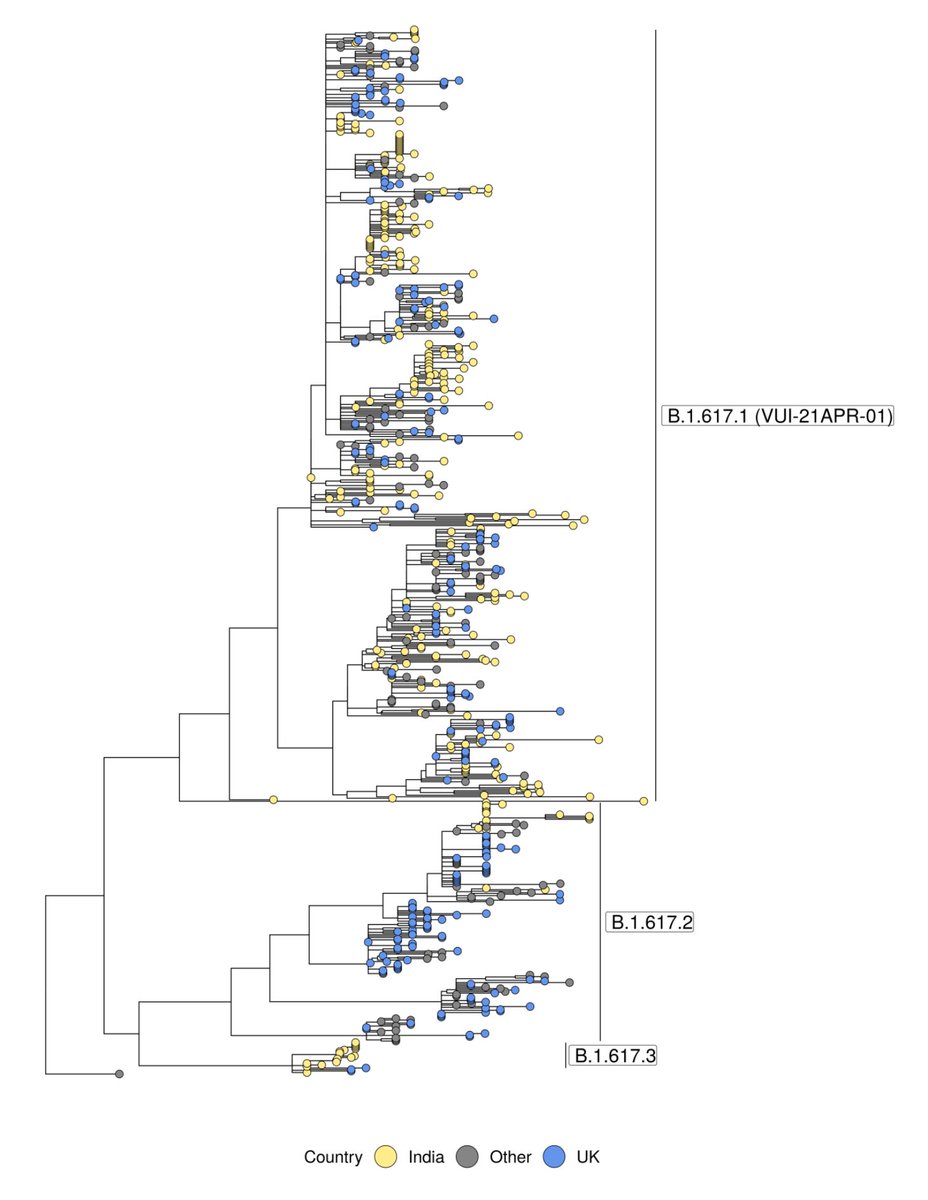
 First the obviously bad stuff (red): nine mutations seen in previous VOCs. There's a lot of overlap already among VOCs (convergent evolution), but this variant has an unprecedented sampling from mutations previously seen in Alpha, Beta, Gamma and Delta separately.
First the obviously bad stuff (red): nine mutations seen in previous VOCs. There's a lot of overlap already among VOCs (convergent evolution), but this variant has an unprecedented sampling from mutations previously seen in Alpha, Beta, Gamma and Delta separately.

 And it has grown all over the place between mid-July (L) and now (R). AY.4 did a similar thing, but it was not displacing other Delta, and given that it hasn't spread through the world, likely just had some epidemiological luck.
And it has grown all over the place between mid-July (L) and now (R). AY.4 did a similar thing, but it was not displacing other Delta, and given that it hasn't spread through the world, likely just had some epidemiological luck. 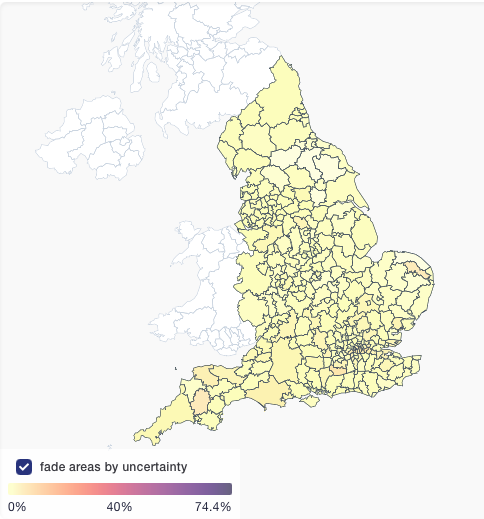


https://twitter.com/babarlelephant/status/1398758089978003462?s=20

https://twitter.com/jcbarret/status/1426461850791723015?s=20

https://twitter.com/gerrythill/status/1427098948729151490This figure shows relative mutation rate: each colour is a type of mutation like C to A, with the bars showing the context of the bases to the left and right; below the line is the reverse complement (e.g. G to T for blue).Red C>U (C>T in DNA) mutations are by far the most common


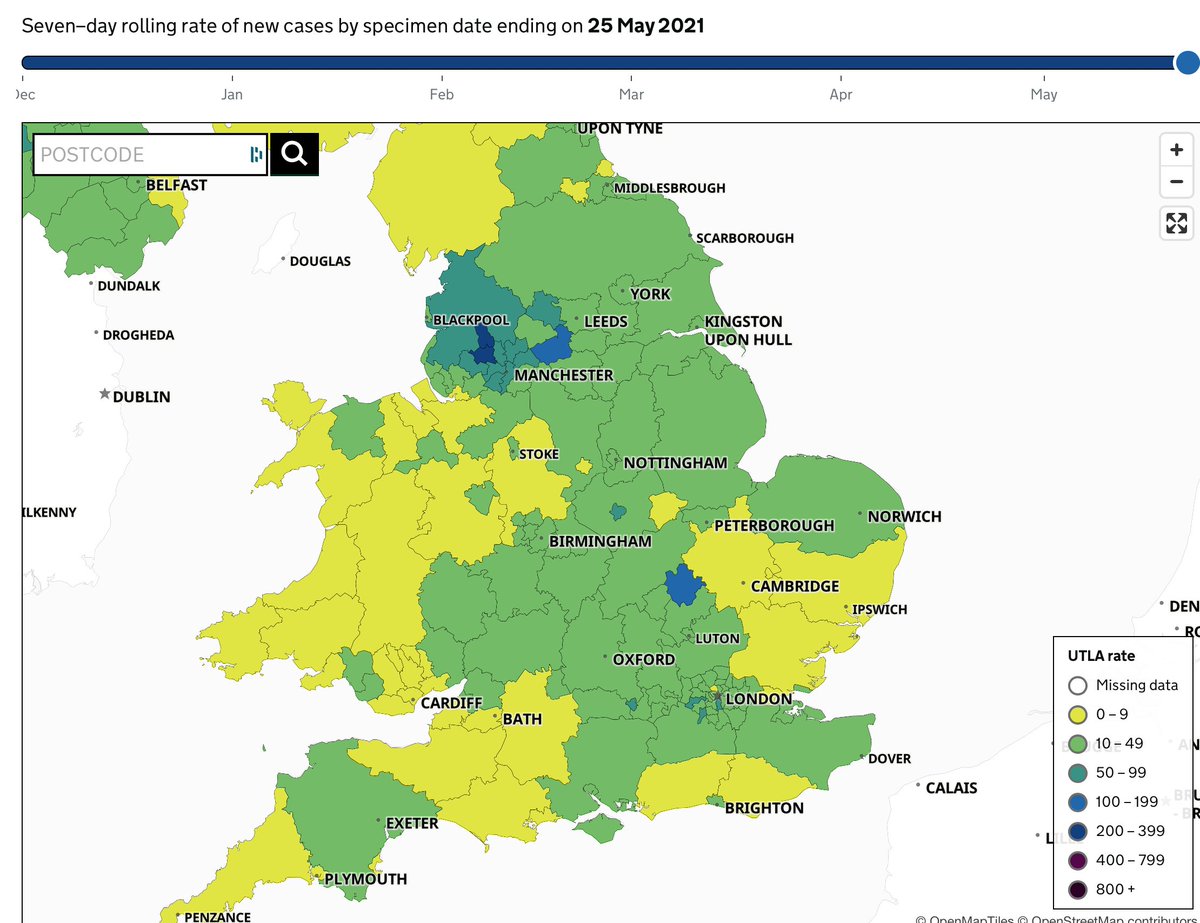
 First new feature is "fade areas by uncertainty", or "nebulosity view", as I like to call it. We fade the colouring of local authorities on proportion view to give a visual hint that you shouldn't over-interpret an area with 100% Delta if it only has a handful of sequences.
First new feature is "fade areas by uncertainty", or "nebulosity view", as I like to call it. We fade the colouring of local authorities on proportion view to give a visual hint that you shouldn't over-interpret an area with 100% Delta if it only has a handful of sequences. 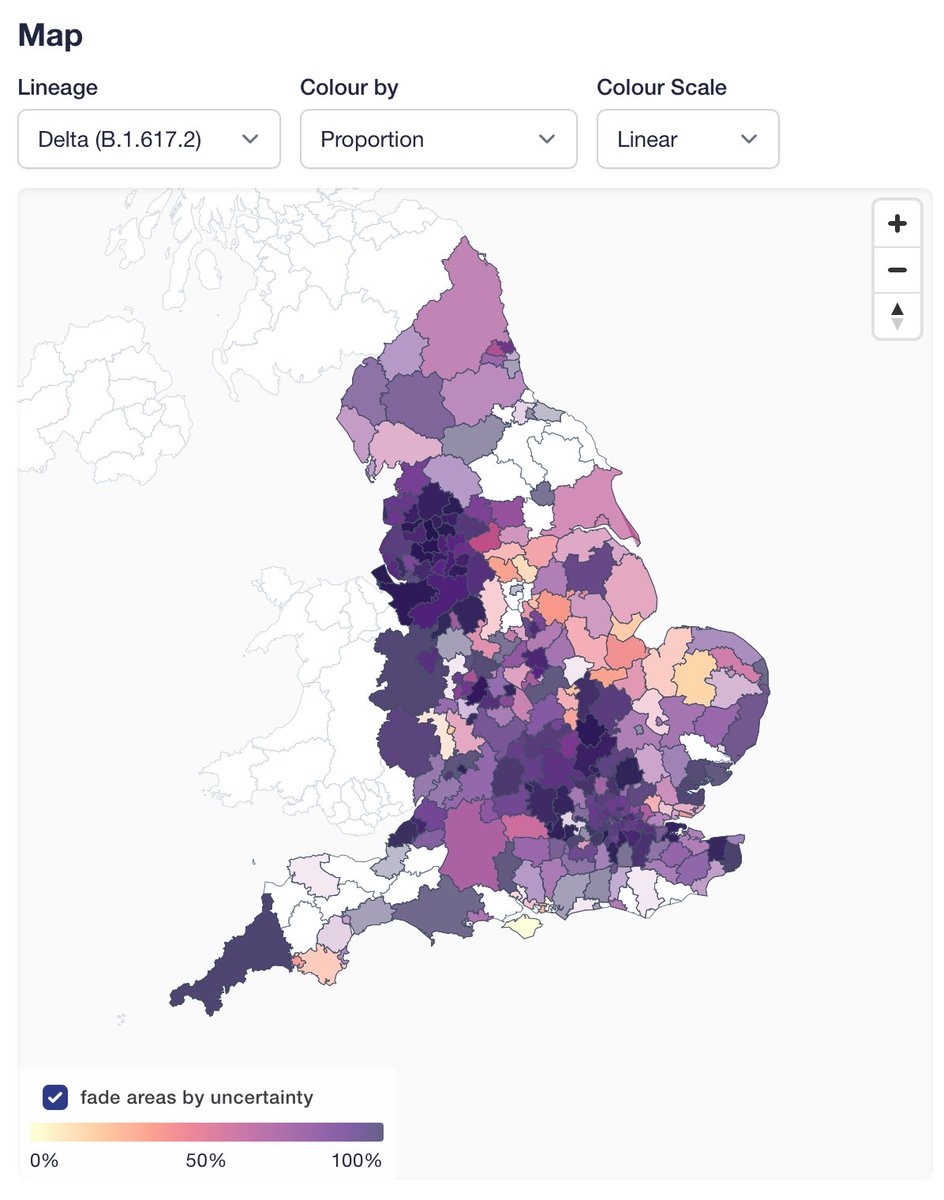



 A couple of notes on the site. We have updated the text describing ascertainment: we are no longer excluding surge tests, which make up a large fraction of all tests in key areas in recent weeks. I think this provides least biased frequencies now. Feedback welcome.
A couple of notes on the site. We have updated the text describing ascertainment: we are no longer excluding surge tests, which make up a large fraction of all tests in key areas in recent weeks. I think this provides least biased frequencies now. Feedback welcome.

https://twitter.com/jburnmurdoch/status/1396531230808625159?s=20&
https://twitter.com/DevanSinha/status/1396575235114209282?s=20

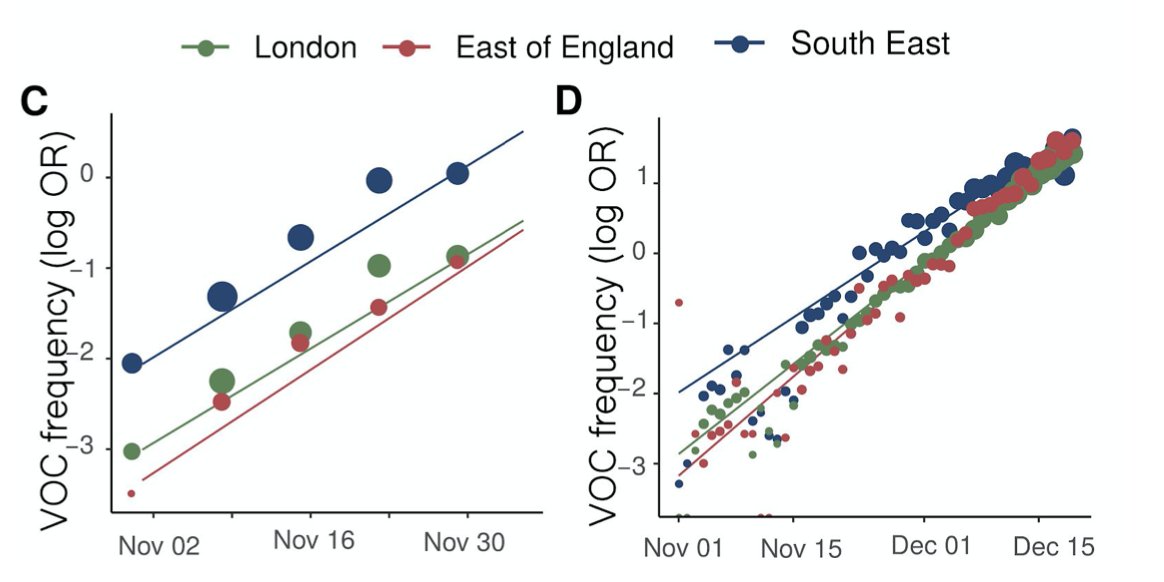
https://twitter.com/kallmemeg/status/1390670671139774467?s=20First, headline result, as reported by others, is that B.1.617.2 has hit 6% in England in our 2-week rolling average (>10% in the most recent week). Other VOCs & VUIs steady.