
i am a biologist/physicist at the @biozentrum in Basel trying to understand how microbes evolve. views are my own, RT!=endorsement.
How to get URL link on X (Twitter) App


 Within each variant, diversity and divergence increase linearly with time. The rate of synonymous evolution is around 6 changes per year in all variants, while the non-synonymous rate varies from variant to variant.
Within each variant, diversity and divergence increase linearly with time. The rate of synonymous evolution is around 6 changes per year in all variants, while the non-synonymous rate varies from variant to variant.

 Since 2017, the lineage leading to the recent samples has a peculiar mutation pattern where almost all mutations are G->A or C->T. Furthermore, they almost all occur in specific sequence contexts as Andrew Rambaut discusses here:
Since 2017, the lineage leading to the recent samples has a peculiar mutation pattern where almost all mutations are G->A or C->T. Furthermore, they almost all occur in specific sequence contexts as Andrew Rambaut discusses here:
 Nick @n_b_noll has developed a PanPraph for scalable construction of pan-genome graphs from closely related bacterial genomes or plasmids. 2/
Nick @n_b_noll has developed a PanPraph for scalable construction of pan-genome graphs from closely related bacterial genomes or plasmids. 2/
 Pierre Barrat has developed am algorithm to infer reassortments between two segments of influenza viruses.
Pierre Barrat has developed am algorithm to infer reassortments between two segments of influenza viruses. 




https://twitter.com/richardneher/status/1293857065425866754
 Comparing the age distributions of confirmed cases in Switzerland before and after June 1st, you see a strong shift towards young adults and very few cases in people >70y.
Comparing the age distributions of confirmed cases in Switzerland before and after June 1st, you see a strong shift towards young adults and very few cases in people >70y. 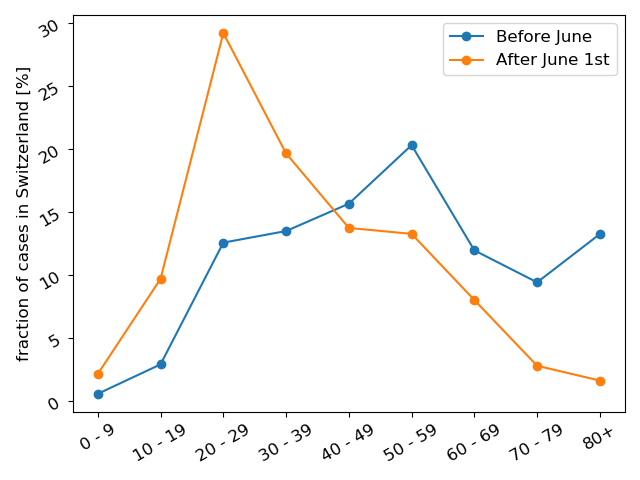

 The 2009 H1N1 influenza pandemic might offer some clues. H1N1pdm is much less deadly, but much like #SARSCoV2 it
The 2009 H1N1 influenza pandemic might offer some clues. H1N1pdm is much less deadly, but much like #SARSCoV2 it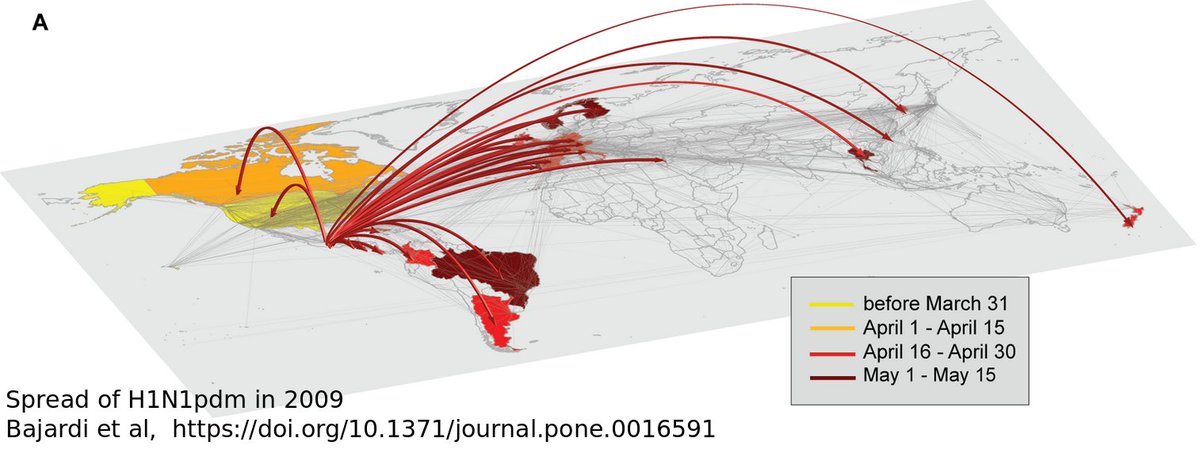

 Pierre found that this is not typical behavior. Mutation trajectories of H3N2 in the last 20y that rapidly rose from 0 to 30% in frequency show no trend of increasing further in frequency. The fixation probability of a mutation at frequency x is almost exactly x! [2/7]
Pierre found that this is not typical behavior. Mutation trajectories of H3N2 in the last 20y that rapidly rose from 0 to 30% in frequency show no trend of increasing further in frequency. The fixation probability of a mutation at frequency x is almost exactly x! [2/7] 

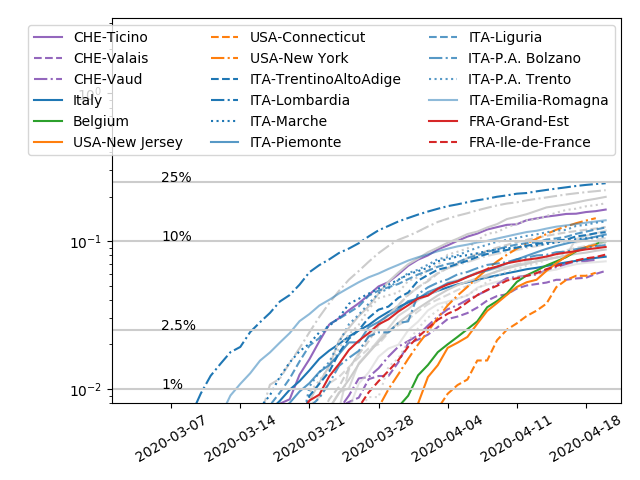
 The graph shows a simple estimate of prevalence assuming an IFR of 0.5% and the same delay to death and seroconversion. The numbers are for countries, states, or regions. There is considerable variation within these and seroprevalence depends a lot on the population tested.
The graph shows a simple estimate of prevalence assuming an IFR of 0.5% and the same delay to death and seroconversion. The numbers are for countries, states, or regions. There is considerable variation within these and seroprevalence depends a lot on the population tested.
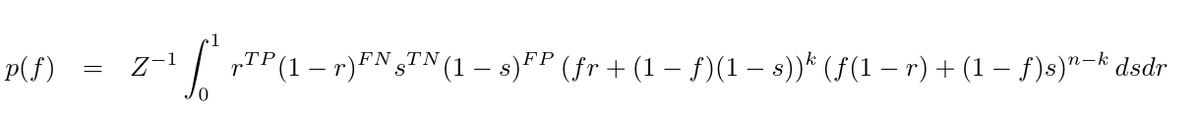
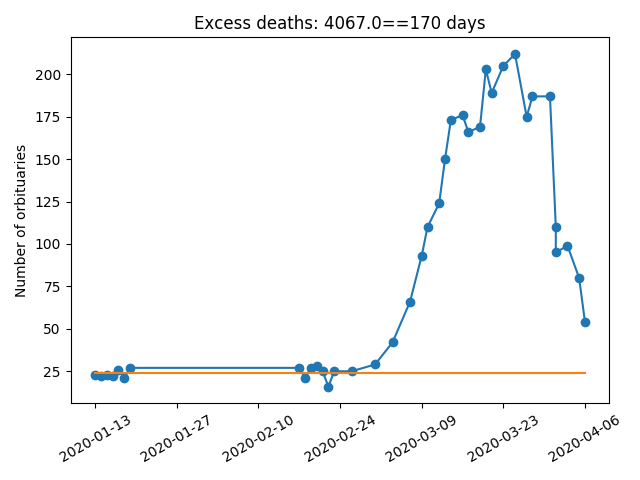

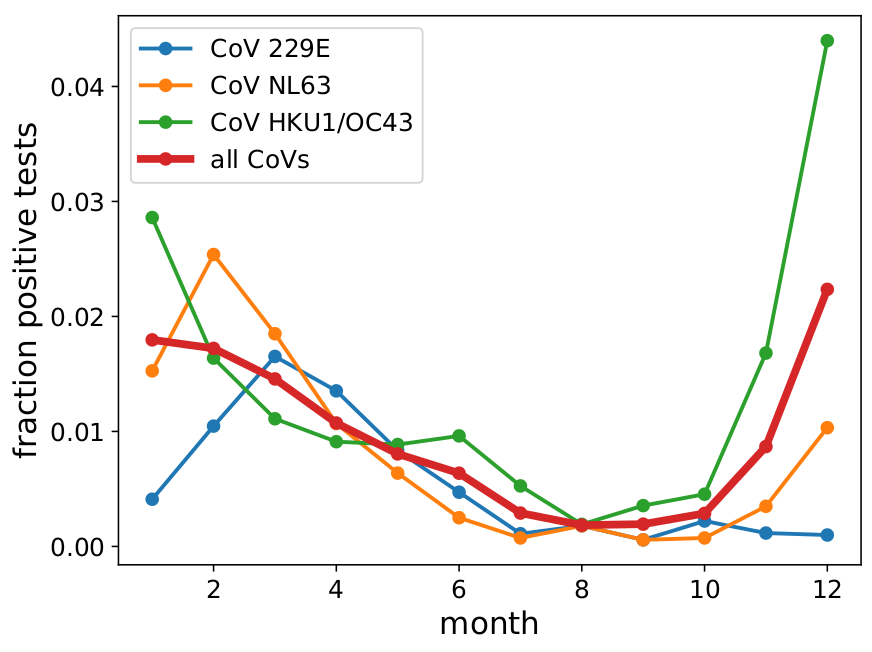

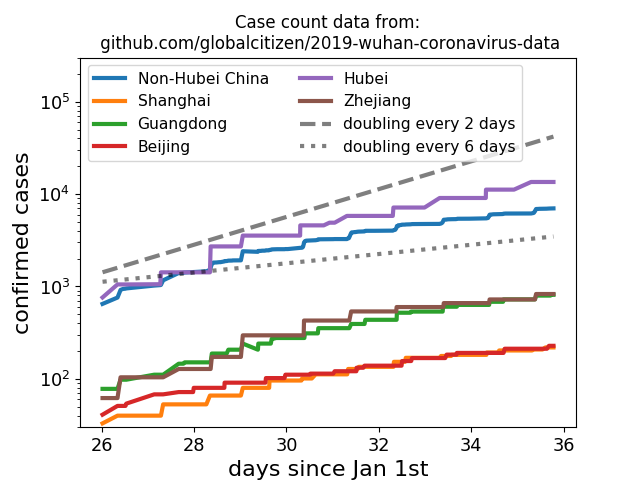
 We don't see a similarly pronounced deceleration in the number of fatalities. Fatalities are expected to lag behind case counts, so this is not unexpected. [2/6]
We don't see a similarly pronounced deceleration in the number of fatalities. Fatalities are expected to lag behind case counts, so this is not unexpected. [2/6] 