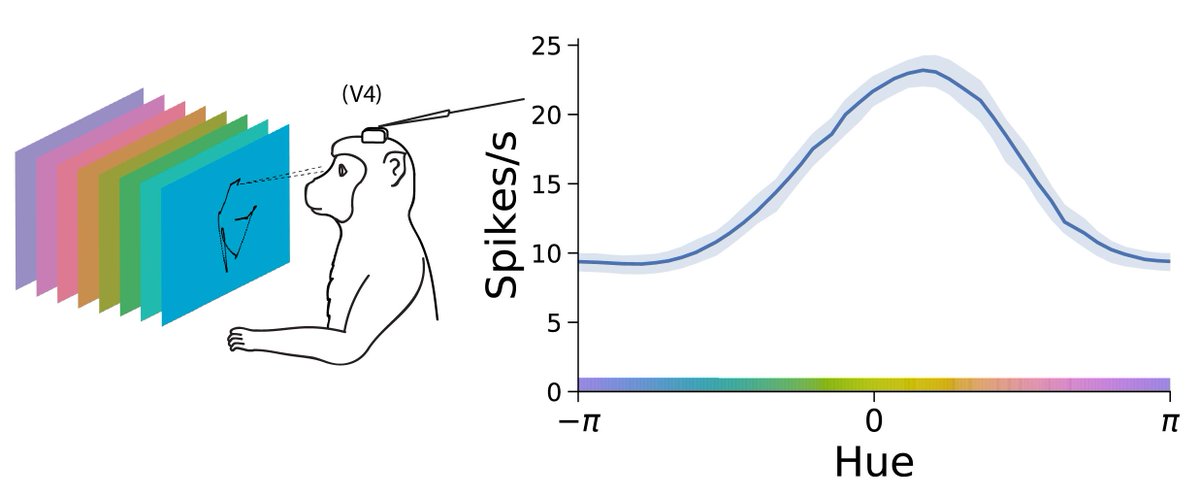
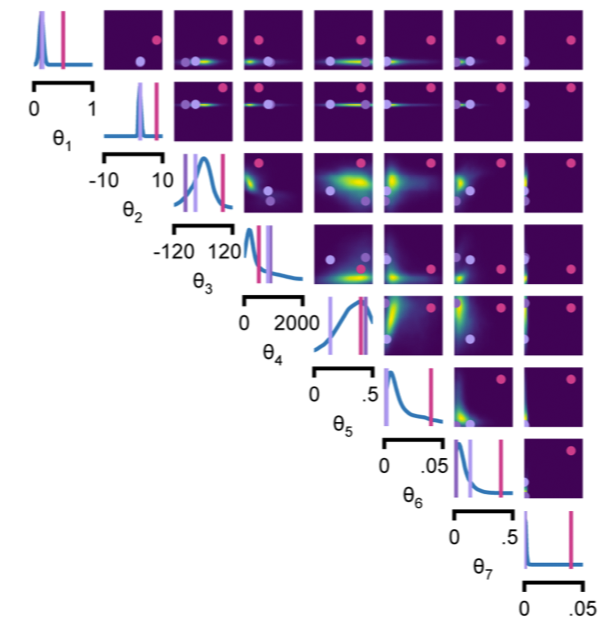
Catch is: Getting models and data to agree can be tricky, and people spend time tuning parameters by hand or with heuristics. (2/n)
Exhibit 1: Ion-channel models (with @TPVogels et al). The approach finds the posterior (read: space of params that work) (6/n)
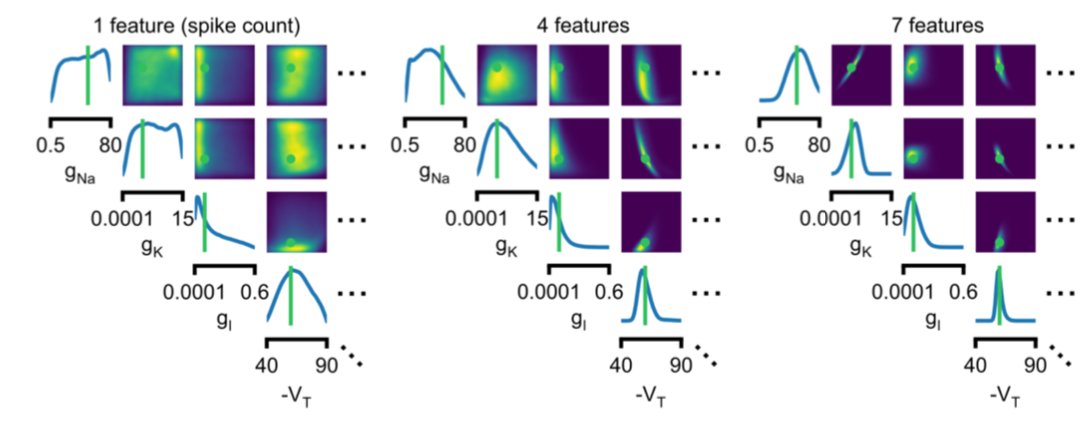
Code mackelab.org/delfi, tutorials still being brushed up.
Many thanks to great co-authors+many others!
@TU_Muenchen @caesarbonn @NNCN_Germany @dfg_public biorxiv.org/content/10.110… END